Shifting of Risk Groups in Acute Myeloid Leukaemia Patients with FLT3–ITD Mutation: Challenge in the Disease Prognosis?
Jayalakshmi T* and Manasa MS
Department of Genetic Engineering, Bharath Institute of Higher Education and Research, India
*Address for Correspondence: Jayalakshmi T, Genetic department, Bharath Institute of Higher Education and Research, Chennai Tamilnadu, India, Tel: +919-003-101-65; E-mail: jayamaniraaja07@gmail.com
Submitted: 09 August 2019; Approved: 03 December 2019; Published: 04 December 2019
Citation this article: Jayalakshmi T, Manasa MS. Shifting of Risk Groups in Acute Myeloid Leukaemia Patients with FLT3–ITD Mutation: Challenge in the Disease Prognosis. Int J Blood Dis Dis. 2019; 3(1): 014-019.
Copyright: © 2019 Jayalakshmi T, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: AML: Acute Myeloid Leukemia; FLT3: FMS-Like Tyrosine Kinase 3; ITD: Internal Tandem Duplication; Prognosis
Download Fulltext PDF
Acute Myeloid Leukemia (AML) is a rapidly growing heterogenous malignancy of blood or bone marrow where the bone marrow produces many abnormal cells called blasts. As majority of AML cases shows chromosomal abnormalities and gene mutations, cytogenetic and molecular analysis are most popular as independent prognostic indicator of AML. Although AML cases can be stratified into favorable, intermediate and adverse-risk groups based on their validated cytogenetics and molecular abnormalities, prognosis within these categories varies widely because of some recurrent gene mutations. Recognizing these genetic changes in patients help to identify certain types of AML and can be important in determining a patient’s individual prognosis and treatment. Internal Tandem Duplication (ITD) of FLT3 (FMS-Like Tyrosine Kinase 3) gene is one of such most common somatic mutations associated with an increased risk of relapse and shorter overall survival unlike recurrent cytogenetic abnormalities such as t(8;21) and inv(16) that promises a better clinical outcome in the majority of cases. Patients with this mutation show a very poor prognosis regardless of their cytogenetic profile that results in the shifting of AML risk stratification and create complications in patient’s treatment and survival rate.
Introduction
Although leukemia is famous as a childhood disease, it is ten times more common in adults accounting for ~80 percent of cases in this group [1]. The American Cancer Society estimates that About 21,450 new cases of Acute Myeloid Leukemia (AML) reports and About 10,920 deaths occur from AML annually. Worldwide, AML reports are highest in the U.S., Australia and Western Europe [2]. Cancer Statistics Review of 2015 by NIH reported that the number of AML deaths was 2.8 per 100,000 men and women per year. Approximately 0.5 percent of men and women will be diagnosed with this abnormality at some point during their lifetime, based on 2013-2015 data. Based on data from SEER 18 2008-2014, only 27.4% of AML patients survive for 5 Years or More after Being Diagnosed with the disease. This shows that acute myeloid leukemia is most frequently diagnosed among people aged 65-74. It has been estimated that the percent of acute myeloid leukemia deaths is highest among people aged 75-84. Studies also show that it is more likely to occur in Caucasian than in African Americans. The reason for this difference still unknown [3].
Mutations in the FMS-Like Tyrosine Kinase 3 (FLT3) gene happens in approximately 30% of all AML cases, either with the Internal Tandem Duplication (ITD) representing the most common type of FLT3 mutation (FLT3-ITD; approximately 25% of all AML cases) or with mutations in the Tyrosine Kinase Domain (TKD) of FLT3 which is less frequent (7%) in AML patients and currently have no clinically significant impact [4]. FLT3-mutated AML is frequently found in patients with cytogenetically normal AML and the prognostic impact of FLT3 mutations has made FLT3 an interesting target [5]. Organization named European Leukemia Net (ELN) proposed a new risk stratification of AML in 2017 by including gene mutations like FLT3-ITD with corresponding allelic ratio that might solve the problem of shifting of risk groups in AML patients with gene mutations. Review.
Normal function of FLT3 gene
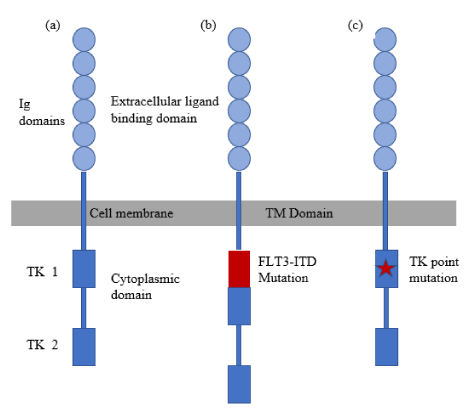
The FMS-Like Tyrosine Kinase 3 (FLT3) gene is a 24-exon gene situated on chromosome 13q12.2, encodes the FLT3 tyrosine kinase receptor expressed on the surface of CD34+ hematopoietic stem cells and other immature hematopoietic progenitors. FLT3 is a member of type-1 transmembrane receptor with five Ig domains in its extracellular domain, transmembrane domain and two kinase domains in its intracellular domain (Figure 1). When the receptor tyrosine kinases bound with a specific protein called FLT3 ligand or FL, corresponding signals are triggered to transmit from cell surface into the cell through a process called signal transduction [6]. It is a class III receptor tyrosine kinase that upon activation promotes the activation of downstream pathways involving Phosphatidylinositol-3 Kinase (PI3K), AKT, Mammalian Target of Rapamycin (mTOR), RAS and extracellular Signal-Related Kinase (ERK). These pathways have an important role in cellular proliferation, survival, and differentiation. In addition to the activation of the receptor by ligands, mutations also constitutively activate the receptor and the cells uncontrolled proliferation [7].
FLT3 protein is 993 amino acids long (112804 Da) and structurally related to the receptors for Platelet Derived Growth Factor (PDGF), Colony Stimulating Factor 1 (CSF1), and KIT Ligand (KL). FLT3 proteins are found to be produced in bone marrow CD34-positive cells, corresponding to multipotential, myeloid and B-lymphoid progenitor cells, and on monocytic cells. In addition, the FLT3 protein is expressed on blast cells from most AML and B-ALL [8]. FLT3 mutations in AML
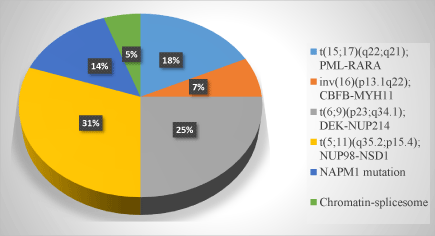
FLT3 gene is frequently overexpressed in approximately 28-30% of AML patients [9]. Figure 2 illustrates the frequency of FLT3 mutations in AML with recurrent genetic abnormalities. Several studies across the world in well-documented AML patients demonstrated that FLT3 mutations are strongly associated with a poor prognosis and a high leukemia cell count in patients with AML, suggesting that FLT3 mutations are involved in disease progression [10]. Although FLT3-mutated Acute Myeloid Leukemia (AML) is not yet recognized as a distinct entity in the World Health Organization (WHO) classification system, it is readily recognized as a challenge by clinical specialists who treat acute leukemia [11]. Mutations in FLT3 gene in AML are classified into the following two types: Internal Tandem Duplications (ITDs) and base substitution mutations in the activation loop of the Tyrosine Kinase Domain (TKDs).
FLT3-TKD mutations
FLT3-TKD mutations are small mutations in the activation loop of tyrosine kinase domain of FLT3, mostly representing point mutations in aspartic acid codon D835 or deletions of codon I836 [12]. Mutations additionally have been observed are I836M, I836T, I836MA and I836ins LK [13]. These mutations induce constitutive tyrosine phosphorylation that leads to conformational change, exposing the active site and resulting in up-regulation of kinase function and ligand-independent activation. However, the effects of TKD mutations in proliferation rate of FLT3 is not as potent as ITDs, the prognostic significance is similar for both mutations [14]. In a meta-analysis conducted by Yanada et al. [15] authors acknowledged that one of the limitations of their study was the difficulty in detecting actual effects on outcome of FLT3-TKD mutations due to its low prevalence. Smith et al. [16] suggested that FLT3-TKD mutations at the D835 codon have been associated with increased rates of resistance to tyrosine kinase inhibitors that target FLT3. A clinical study of 2502 AML patients done in 2006 found a relatively high frequency of FLT3-TKD in t(15;17)/ PML-RARA compared with the total cohort and suggested that FLT3-TKD significantly associated with the FAB subtype M3v (11.8%), whereas in M3 the frequency (4.7%) was comparable to the overall frequency of FLT3-TKD mutations in AML [17].
Internal tandem duplications of FLT3 (FLT3-ITDS)
Internal Tandem Duplication (ITD) mutation in the FLT3 gene are common in AML (20% of all cases), especially in normal karyotype disease (28-34%), where they are found commonly to co-occur with NPM1 mutations and are associated with an inferior outcome [18]. Mutations most commonly occur in sequences of exons 14 and 15 of the FLT3 gene and show a large variation in the number and sizes of duplicated fragments [19]. Mutations results in constitutive activation of the receptor tyrosine kinase and downstream activation of RAS, MAPK and STAT5 signalling pathways leading to dysregulated cellular proliferation. FLT3-ITDs represent an unfavourable prognosis and inferior overall survival due to a high relapse rate. AML patients with FLT3-ITD mutation achieve complete remission rates comparable to those of patients with wild-type disease but have significantly higher rates of relapse and shorter durations of disease-free and Overall Survival (OS) [20]. Researches on FLT3-ITD mutations has revealed a challenging crisis in treatment of AML patients with this mutation. That is FLT3-ITD-expressing human myeloid cell lines are resistant to cytosine arabinoside, which is a most common drug used for consolidation therapy in adult AML patients. Additional, in vitro evidence also suggests that FLT3-ITD mutations may act as a key contributor to this drug resistance [21]. This has led to intensive research to identify novel therapeutic inhibitors for FLT3 mutations.
Prognostic significance of FLT3-ITD in AML
Patients with FLT3-ITD mutation show leukocytosis, high blast counts, increased risk of relapse, shorter overall survival, normal cytogenetics, t(15;17), t(6;9), NPM1 and DNMT3A mutations [22]. Regarding AML FAB subtypes, FLT3 mutations are clearly more common in the M3 and M4 subtypes followed by M2, M1 and M5b subtypes [23]. Interestingly, FLT3-ITD mutation is rare in M6 and M7 subtypes [24]. Since most of the AML cases with this mutation confers poor prognosis, it creates a significant negative impact on the management of patients with AML [25]. Clinical studies show a significant increase in the number of blasts in AML patients with FLT3-ITD mutation at relapse than at diagnosis, suggesting that FLT3-ITD may function as the driver mutation responsible for the progression of disease and drug resistance [26]. As FLT3-ITD mutations most commonly found in the intermediate cytogenetic risk group especially in normal karyotype AML patients, the impact of FLT3 ITD mutation status at relapse is expected to be of greater prognostic value than the mutation status at diagnosis in normal karyotype-AML patients with relapse [27].
Clinically validated technique for FLT3-ITD mutation detection in AML patients is Polymerase Chain Reaction (PCR) amplification of wild type and mutant alleles isolated from a sample of peripheral blood or bone marrow, followed by agarose-gel electrophoresis and ITD size determination by sequencing analysis. In this way, amplicons larger than those of the wild type are interpreted as positive for ITD mutation [28]. In addition to ITD length determination, Mutant-to-wild-type allelic ratio, insertion site, karyotype, and the presence of a mutation in the NPM1 gene appear to have further influence in the prognostic utility of FLT3-ITD in patients with newly diagnosed FLT3-ITDmutated AML [29]. There are many clinical studies, those prove the significance of mutant-to-wild-type allelic ratio in AML prognosis and survival rate. The FLT3 mutant-to-wild type allelic ratio simply means the fraction of leukemia cells that harbor the mutation [30]. A FLT3 Internal Tandem Duplication (ITD) is regarded as unfavorable only if the allelic ratio has a threshold of > 0.5.18 [31]. Prognostic relevance of FLT3-ITD in association with Nucleophosmin-1 (NPM1) mutation is currently a matter of debate [32]. Recent studies reported a better outcome in AML patients with FLT3-ITD low allelic ratio ( < 0.5) in the presence of Mutant Nucleophosmin-1 (NPM1) whereas this protective effect of NPM1 in AML with higher FLT3-ITD allelic ratio ( ≥ 0.50) is diminished or gets completely lost and was moved into the high-risk group [33].
Discussion
The large subset of AML comprises approximately 40% to 50% of patients with normal karyotype. Although such cytogenetically normal AML cases are currently under intermediate-risk group, it is quite heterogeneous, and not all patients in this subset have the same response to treatment. This may be a result of variations in gene mutations like FLT3-ITD among people [34]. Characterization of chromosomal abnormalities and gene mutations in AML helps in the stratification of patients according to risk and guide therapeutic decisions. According to guidelines established by several organizations, including the World Health Organization (WHO), National Comprehensive Cancer Network (NCCN), and European Leukemia Net (ELN), AML cases are classified into mainly three risk groups, favorable, Intermediate and poor based on the presence of certain cytogenetic and molecular aberrations [35]. Although WHO guidelines, published in 2008 (Table 1) listed FLT3-ITD in favorable and poor risk group with specific cytogenetic subgroups, it does not group FLT3 mutations into a single category and thus had limitations in the prognosis of AML patients with FLT3-ITD [36]. The NCCN Clinical Practice Guidelines in 2017 classify patients with AML with normal cytogenetics harboring the FLT3-ITD or TP53 mutations as poor risk and those with NPM1 mutation in the absence of FLT3-ITD as favorable risk. ELN put forward an advanced risk classification of AML in 2017 along with FLT3-ITD screening, mutant to-wild-type allelic ratio and Tyrosine Kinase Domain (TKD) mutations at codons D835 and I836. This detailed ELN recommendations were found to valid in a large retrospective analysis in patients with newly diagnosed AML and intermediate-risk cytogenetic abnormalities or normal karyotype [37]. Overall, it is clear from the table 1 that, occurrence of FLT3-ITD mutation in AML cases shift AML risk groups based on mainly three criteria such as presence of NAPM1 mutation and FLT3-ITD allelic ratio.
Clinical significance of FLT3 mutations from diagnosis to relapse suggest that testing for FLT3-ITD mutations may be necessary at multiple time points throughout a patient’s disease course to help guide the most appropriate therapeutic decisions like targeted treatments that may help patients achieve longer and more durable remissions. Although ELN risk stratification is appeared to be very useful, there are still some problems associated with this. The main crisis relies on FLT3-ITD allelic ratio data, which has yet to become part of the standard testing in clinical practice with an internationally standardized methodology and is often unavailable to treating physicians [38]. Moreover, interpretation of test results is often difficult due to variability in diagnostic accuracy, sensitivity, and qualitative vs. quantitative readouts of different FLT3 assays [39]. Once researchers could solve these problems and able to standardize values for FLT3-ITD allelic ratios corresponding to AML risk status, an advanced AML risk classification could be developed with more accurate data beyond what is already described by ELN 2017 classification.
Considering the high frequency with which FLT3 mutations occur in AML, research has been going on worldwide in order to develop FLT3 inhibitors. A number of TKIs (Tyrosine Kinase Inhibitors) are under development that disrupt the oncogenic signaling initiated by FLT3 along with a variety of improved treatment strategies in AML. The recognition that FLT3-ITD is an adverse prognostic marker, the integration of FLT3 inhibitors into the treatment algorithm, and the increased use of Allogeneic Hematopoietic stem cell transplantation have led to improvements over the past 15 years in clinical outcomes in patients with FLT3-ITD-mutated AML [40].
Midostaurin, one of the three first generation multi-kinase inhibitors (Sorafenib, Lestaurtinib, and Midostaurin) represent the first FDA approved FLT3 inhibitor for the treatment of patients with FLT3-mutated AML. The failure of these agents to induce durable responses led to the development of second generation FLT3 tyrosine kinase inhibitors (Quizartinib, Crenolinib, Gilteritinib) those overcome the resistance to first-generation agents [41] (Table 2). Although Quizartinib demonstrated favorable outcomes in clinical studies recently, acquired clinical resistance to Quizartinib has been an emerging issue in treatment [42]. To overcome these problems, a novel FLT3inhibitor, FF-10101 has been designed with selective and irreversible FLT3 inhibition [42]. Since clinical trials confer the potency of FF-10101 is higher among the developed FLT3 inhibitors, researchers highly expect better clinical efficacy in patients with AML with FLT3 mutations than those of previously developed FLT3 inhibitors [43].
Conclusion
As both the NCCN and ELN 2017 guidelines for AML risk stratification support FLT3 testing for all patients with AML, it solves the problem of shifting of risk groups in AML cases with FLT3-ITD mutations to a great extend allowing better understanding of the patient’s disease and enable to take most appropriate therapeutic decisions. However, there is currently no internationally standardized methodology for determining these allelic ratios create a challenge in AML prognosis that want to be rectified immediately.
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016; 127: 2391-2405. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27069254
- Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: A study of 2502 patients. Blood. 2006; 107: 3847-3853. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/16434492
- Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S. Prognostic relevance of FLT3-TKD mutations in AML: The combination matters-an analysis of 3082 patients. Blood. 2008; 111: 2527-2537. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/17965322
- Badar T, Kantarjian HM, Nogueras GGM, Borthakur G, Garcia Manero G, Andreeff M, et al. Improvement in clinical outcome of FLT3 ITD mutated acute myeloid leukemia patients over the last one and a half decade. Am J Hematol. 2015; 90: 1065-1070. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/26299958
- Chen Y, Pan Y, Guo Y, Zhao W, Ho WT, Wang J, et al. Tyrosine kinase inhibitors targeting FLT3 in the treatment of acute myeloid leukemia. Stem Cell Investig. 2017. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/28607922
- Craig AA, Sheila AB. Cancer: Basic Science and Clinical Aspects. United Kingdom; Wiley-Blackwell publication: 2010. 418. http://bit.ly/2DGG1AO
- Damiani D, Mario Tiribelli. Molecular landscape in adult acute myeloid leukemia: Where we are where we going?". JLPM. 2019; 4. http://bit.ly/360BsgM
- Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia. 2019; 33: 299-312. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30651634
- De Kouchkovsky I. Abdul-Hay M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016; 6: e441. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27367478
- Deschler B, Lubbert M. Acute myeloid leukemia: Epidemiology and etiology. Cancer. 2006; 107: 2099-2107. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/17019734
- Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017; 129: 424-447. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27895058
- El Fakih R, Rasheed W, Hawsawi Y, Alsermani M, Hassanein M. Targeting FLT3 mutations in acute myeloid leukemia. Cells. 2018; 7: 4. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/29316714
- Elshoury A, Przespolewski A, Baron J, Wang ES. Advancing treatment of acute myeloid leukemia: The future of FLT3 inhibitors. Expert Rev Anticancer Ther. 2019; 19: 273-286. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30681373
- Estey EH. Acute myeloid leukemia: 2019 update on risk‐stratification and management. Am J Hematol. 2018; 93: 1267-1291. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30328165
- Grafone T, Palmisano M, Nicci C, Storti S. An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: Biology and treatment. Oncol Rev. 2012; 6: e8. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25992210
- Hannum C, Culpepper J, Campbell D, McClanahan T, Zurawski S, Bazan JF, et al. Ligand for FLT3/FLK2 receptor tyrosine kinase regulates growth of haematopoietic stem cells and is encoded by variant RNAs. Nature. 1994; 368: 643-648. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/8145851
- Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, et al. SEER Cancer Statistics Review (CSR) 1975-2016. National Cancer Institute. http://bit.ly/2Yh4ZAj
- Kayser S, Levis MJ. FLT3 tyrosine kinase inhibitors in acute myeloid leukemia: Clinical implications and limitations. Leuk Lymphoma. 2014; 55: 243-255. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/23631653/
- Kiyoi H, Naoe T. FLT3 mutations in acute myeloid leukemia. Methods Mol Med. 2006; 125: 189-197. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/16502586
- Ko YC, Hu CY, Liu ZH, Tien HF, Ou DL, Chien HF, et al. Cytarabine-resistant FLT3-ITD leukemia cells are associated with TP53 mutation and multiple pathway alterations-possible therapeutic efficacy of cabozantinib. Int J Mol Sci. 2019; 20. 1230. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30862120
- Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, et al. The presence of a FLT3 internal tandem duplication in patients with Acute Myeloid Leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001; 98: 1752-1759. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/11535508
- Kumar Chandra. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer. 2011; 2: 95-107. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/21779483/
- Lagunas-Rangel FA, Chavez-Valencia V. FLT3-ITD and its current role in acute myeloid leukaemia. Med Oncol. 2017; 34: 114. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/28470536
- Levis M. FLT3 mutations in acute myeloid leukemia: What is the best approach in 2013?. Hematology Am Soc Hematol Educ Program. 2013; 2013: 220-226. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/24319184
- Meshinchi S, Alonzo TA, Stirewalt DL, Zwaan M, Zimmerman M, Reinhardt D, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006; 108: 3654-3661. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/16912228/
- Mrozek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012; 30: 4515-4523. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/22987078/
- Murphy KM, Levis M, Hafez MJ, Geiger T, Cooper LC, Smith BD, et al. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003; 5: 96-102. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12707374/
- O'Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Arber DA, et al. Acute myeloid leukemia, version 3.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017; 15: 926-957. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/28687581
- Park SH, Chi HS, Min SK, Cho YU, Jang S, Park CJ, et al. Prognostic significance of the FLT3 ITD mutation in patients with normal-karyotype acute myeloid leukemia in relapse. Korean J Hematol. 2011; 46: 88-95. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/21747880
- Pratz KW, Levis M. How I treat FLT3-mutated AML. Blood. 2017; 129: 565-571. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27872057
- Sabrina Tosi Alistair G. Reid. The genetic basis of haematological cancers. United Kingdom; John Wiley & Sons Ltd publication: 2016. http://bit.ly/2Rj5c4s
- Johannes S, Christoph R, Sabine K, Friedrich S, Kerstin SE, Mathias Haenel, et al. Validation of the ELN 2017 classification for AML with intermediate risk cytogenetics with or without NPM1-mutations and high or low ratio FLT3-ITDs. Blood. 2017; 130: 2694. http://bit.ly/33PgiRl
- Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, et al. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014; 124: 3441-3449. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25270908
- Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: Correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002; 100: 59-66. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12070009
- Smith CC, Lin K, Stecula A, Sali A, Shah NP. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia. 2015; 29: 2390-2392. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/26108694
- Stanicka J, Russell EG, Woolley JF, Cotter TG. NADPH oxidase-generated hydrogen peroxide induces DNA damage in mutant FLT3-expressing leukemia cells. J Biol Chem. 2015; 290: 9348-9361. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25697362
- Tallman MS, Wang ES, Altman JK, Appelbaum FR, Bhatt VR, Bixby D, et al. Acute myeloid leukemia, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019; 17: 721-749. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/31200351
- Thomas CM, Campbell P. FLT3 inhibitors in acute myeloid leukemia: Current and future. J Oncol Pharm Pract. 2019; 25: 163-171. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30270754
- Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002; 100: 2292-2302. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12239137
- Wu, Mei, Chuntuan Li, Xiongpeng Zhu. FLT3 inhibitors in acute myeloid leukemia. J Hematol Oncol. 2018; 11: 133. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30514344
- Yamaura T, Nakatani T, Uda K, Ogura H, Shin W, Kurokawa N, et al. A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells with FLT3 mutations. Blood. 2018; 131: 426-438. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/29187377
- Yanada M, Matsuo K, Suzuki T, Kiyoi H, Naoe T. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: A meta-analysis. Leukemia. 2005; 19: 1345-1349. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/15959528
- Short NJ, Kantarjian H, Ravandi F, Daver N. Emerging treatment paradigms with FLT3 inhibitors in acute myeloid leukemia. Ther Adv Hematol. 2019; 10. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/30800259/

Sign up for Article Alerts