A Multiple Sequence Alignment and in Silico Analysis of GLT4 Protein?
Ibtihal H. Badawi1, Salwa S. Bawazir1, Mohamed S. El-Naggar2 and Hend M. Tag1,2*
1Department of Biology, College of Science and Arts at Khulis, University of Jeddah, Saudi Arabia
2Department of Zoology, Faculty of Sciences, Suez Canal University, Ismailia, 41522, Egypt
*Address for Correspondence:Hend Maarof Tag, Department of Biology, College of Science and Arts at Khulis, University of Jeddah, Saudi Arabia, ORCID: http://orcid.org/0000-0001-9417-1844; E-mail: hend_taha@science.suez.edu.eg
Submitted: 06 August 2020; Approved: 17 August 2020; Published: 20 August 2020
Citation this article: Hansen Badawi IH, Bawazir SS, El-Naggar MS, Tag HM. A Multiple Sequence Alignment and in Silico Analysis of GLT4 Protein. Int J Pharma Anal Acta. 2020;3(1): 025-029.
Copyright: © 2020 Hansen Badawi IH, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: GLT4 protein; Animal model; Human diabetes; Sequence alignment
Download Fulltext PDF
Experimental animals often act as a model of an application similar to humans in most biological processes. However, animal models do not adequately represent an integrated model of similar biological processes) for example, the precise genetic and phenotypic characteristics of human disease). Such systems that operate with the same mechanics can be found in humans and can be treated in the same way in the case of human diseases. Comparison of diseases experienced by humans is similar to that of experimental animals. This study aims to use molecular biology to identify suitable animal models. GLUT4 is an insulin-regulating glucose carrier primarily found in adipose tissue of the striated muscle and tissue (skeleton and heart). This study suggests that because of the high similarity between human and Rattus norvegicus as well as Mus musculus GLUT4, they can be used as a proper animal model for some kind of human diabetes.
Introduction
Multiple sequence alignments denote the basics for comparative analysis designed for identification and description of functional elements. For example, resemblance across large evolutionary distances, detected by a multiple alignment of homologous sequences from several species, usually reveals conserved, and by implication, important, biological features [1,2]. The new technology for full genome sequencing employed after the year 2000 led to accumulation in the GenBank of ClustalW implements an advanced method for multiple sequence alignment and is a widely used tool for DNA or proteins [3]. It calculates the best match for the selected sequences, and lines them up so that the identities, similarities and differences can be found. The algorithm ClustalW proceeds in three steps: pairwise alignment, guide tree and multiple alignments [4].
The Basic Local Alignment Search Tool [BLAST] is the most widely used sequence alignments program [5]. The present work designed to use the previous tools for determining the best type of experimental animal that is most analogous to humans to conduct studies to improve the work of these receptors in the case of diabetes.
Glucose is the major energy and carbon source in mammalian cells. Insulin plays a key role in the whole-body glucose homeostasis, primarily by stimulating glucose uptake into adipose and skeletal muscle tissues [6]. Movement of glucose across plasma membranes is dependent upon either sodium-dependent or facilitative glucose transporters [7]. Facilitative glucose transporters [GLUTs], which utilize the diffusion gradient across the plasma membrane, are ubiquitous [8]. GLUT4 plays a pivotal role in whole-body glucose homeostasis, mediating the uptake of glucose-regulated by insulin [9].
The present study aims to determine the type of experimental animal that is most analogous to humans to conduct studies to improve the work of these receptors in the case of diabetes through serial alignment using in silico Approaches.
Materials and Methods
Sequences, alignment, and construction of phylogenetic tree
Amino acids and the nucleotide sequences for the GLUT4 protein of 12 vertebrate species as well as Homo sapiens were taken from the National Center for Biotechnology Information database [http://www.ncbi.nlm.nih.gov]. The accession numbers of the corresponding database entries and species names are listed in Table 1. The phylogenetic tree was constructed based on the selected reference species. Sequences were multiply aligned in Molecular Evolutionary Genetics Analysis [MEGA] software version 10.1.8 using the Clustal W algorithm and trimmed manually where necessary. Phylogenetic trees were constructed with neighbour-joining [10] and maximum likelihood [11] algorithms using MEGA [12]. The stability of the topology of the phylogenetic tree was assessed using the bootstrap method with 100 repetitions [13]. A distance matrix was generated using the Tamura 3-parameter model [14,15]. All positions containing gaps and missing data were eliminated from the dataset [complete deletion option] using G blocks software.
Results
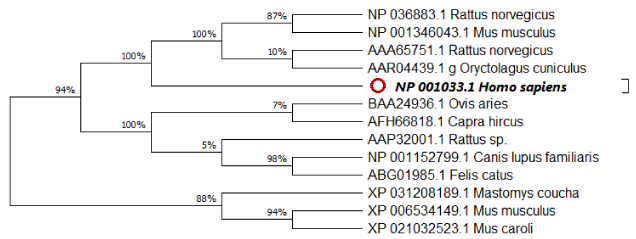
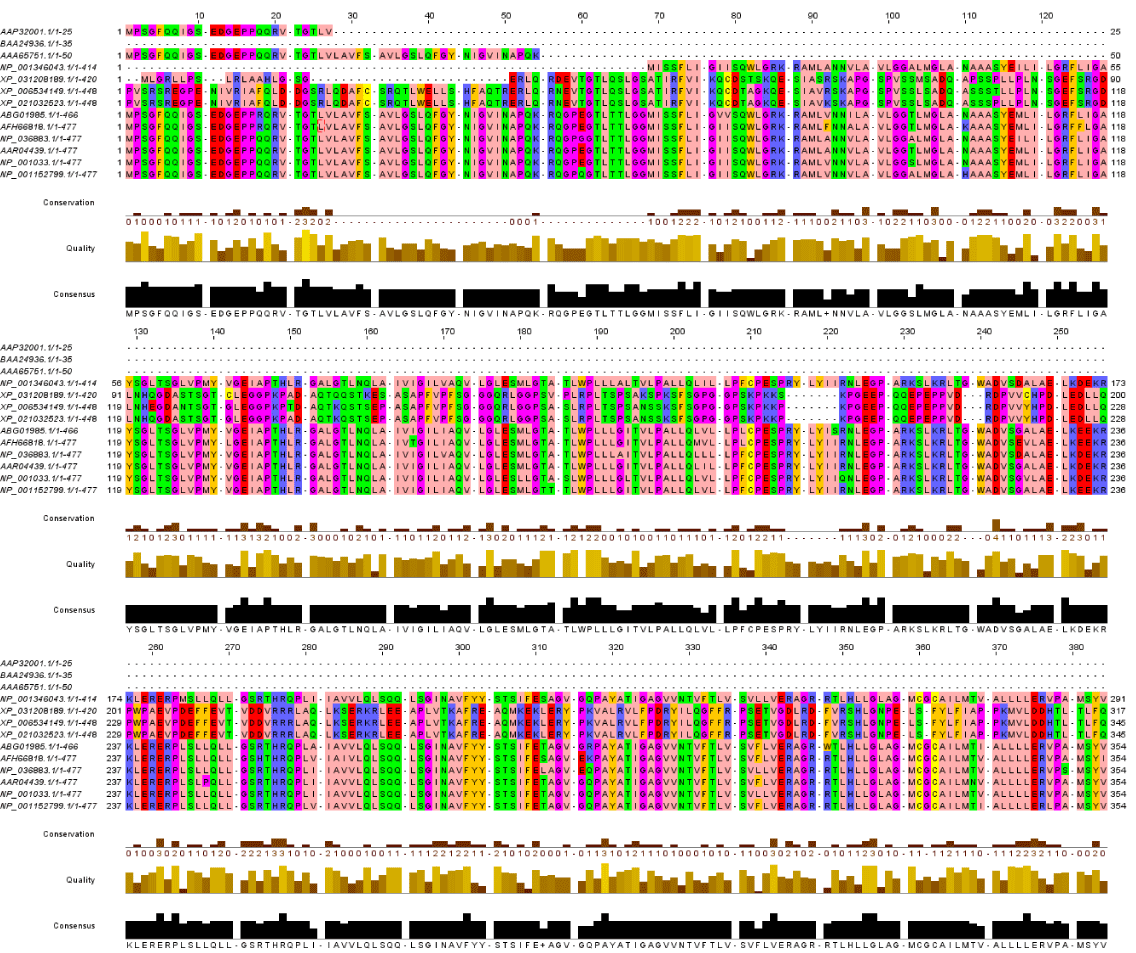
Regarding Phylogeny estimation for GLUT 4 from Homo sapiens and other 12 vertebrate’s species. Neighbour-joining tree constructed using Mega 10.1.8. The bootstrap method used for test of phylogeny was Jones-Taylor-Thornton for detection of the close-neighbour-interchange on random trees. The multiple sequence alignment of the GLUT 4 proteins are depicted in Figure2 respectively. Constructed phylogenetic tree is depicted in figure 1.
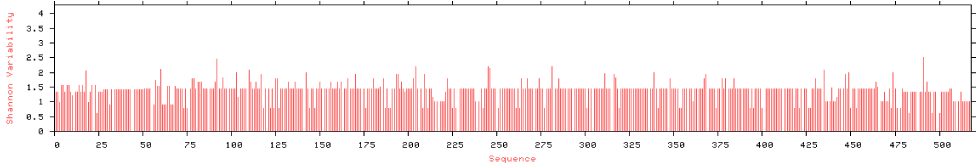
The current results indicated that; Neighbour-joining tree of GLUT 4 displays that there is a high resemblance between human and Mus musculus [NP_001346043.1] as well as Rattus norvegicus [NP_036883.1]. Moreover, figure 3 represents the scoring of each amino acids according to frequent occurring in the sequence of the original alignment; the results indicating that many amino acid has been conserved along alignments which indicating resemblance between human and tested vertebrate’s species. As displayed in figure 4 entropy plot indicates variation in different positions of multiple sequence alignment. Moreover, entropy increases by increasing variation Protein were used for determining entropy plot of GLUT 4 protein.
Discussion
Phylogenetic analysis is a standard and essential tool in any molecular biologist’s bioinformatics toolkit that, in the context of protein sequence analysis, enables us to study the evolutionary history and change of proteins and their function. Such analysis is essential to understanding major evolutionary questions, such as the origins and history of macromolecules, developmental mechanisms, phenotypes, and life itself. On a more practical level, phylogenetic analysis of protein sequence data is integral to gene annotation, prediction of gene function, the identification and construction of gene families, and gene discovery [16].
We used RAxML version 8.0 [17] to infer phylogenetic relationships among the samples. The RAXML (Randomized Axelerated Maximum Likelihood) program has been developed to perform both sequential [on a single processor] and parallel (on multiple processors) phylogenetic analysis using the maximum likelihood optimality criterion [18]. In the present study, the models of evolution used were Jones-Taylor-Thornton (JTT) as selected in Model Test (MEGA X) [15].
Based on accession numbers provided in Table 1, there are repetitive and partial sequences available for some species. Alignment results show that there are four conserved amino acids (F, L, G, M, N, P, W, and V) in all of GLUT 4 sequences. Furthermore, human GLUT4 and R. norvegicus have more amino acids. It shows that these sequences have additional exon on messenger RNA [mRNA] sequence of this gene, or maybe mRNA splicing of these sequences performs in different manners [19]. The most similar sequence to human GLUT4 is Mus musculus. Our finding was in agreement with [20] who reported that functional proteins are known to contain stretches of amino acid sequences highly conserved in different protein families and across species. These conserved sequences constitute protein domains that are generally integral structural units, conferring specific functionalities and often self-folding. The observation of highly conserved protein domains dispersed across different families and organisms provoked the question of whether the domains were reused in some way during evolution.
Calculating Sequence Harmony for GLUT4 from Homo sapiens against 12 sequences was divided into two groups. Group1 represents AFH66818: Capra hircus; AAA65751: Rattus norvegicus; AAP32001: Rattus sp_031208189: Mastomys coucha; XP_021032523: Mus caroli; XP_006534149: Mus musculus and BAA24936: Ovis Aries. While group 2 were consists of 4 species and considered as most related to NP_001033 Homo sapiens; as follows: NP_001346043: Mus musculus; NP_036883: Rattus norvegicus; AAR04439: Oryctolagus cuniculus; NP_001152799 Canis lupus and ABG01985 Felis catus.
So, this study suggests that because of the high similarity between human and Rattus norvegicus as well as Mus musculus GLUT4, they can be used as a proper animal model for some kind of human diabetes. Also, the evolutional changes of GLUT4 are probably caused by point mutations and same exons expressed in this protein sequence because the length of the three integrated sequences are similar and just one amino acid deletion exists in the sequence of Cavia porcellus. Finally, following the changes, which are caused by point mutations, Mus caroli GLUT4 appeared.
Conclusion
The amino acids F, L and P are three conserved and probably key amino acids in GLUT4 sequences which are conserved in Rattus norvegicus as well as Mus musculus, and considered as the most related species to human. It is probable that the position of these amino acids in three-dimension construction of GLUT4 is highly preserved and have a significant role in its function.
- Michael Brudno, Alexander Poliakov, Asaf Salamov, Gregory M Cooper, Arend Sidow, Edward M Rubin, et al. Automated whole-genome multiple alignment of rat, mouse, and human. Genome Res. 2004; 14: 685-692. DOI: 10.1101/gr.2067704
- Berthold Göttgens, Linda M Barton, Michael A Chapman, Angus M Sinclair, Bjarne Knudsen, Darren Grafham, et al. Transcriptional regulation of the stem cell leukemia gene [SCL]-comparative analysis of five vertebrate SCL loci. Genome Res. 2002; 12: 749-759. DOI: 10.1101/gr.45502
- Chaichoompu K, Kittitornkun S, Tongsima S. MT-ClustalW: Multithreading multiple sequence alignment. In: Proceedings 20th IEEE International Parallel & Distributed Processing Symposium. 2006. p. 8. DOI: 10.1109/IPDPS.2006.1639537
- Kuo-Bin Li. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics. 2003; 19: 1585-1586. DOI: 10.1093/bioinformatics/btg192
- David W Mount. Using the basic local alignment search tool [BLAST]. Cold Spring Harb Protoc. 2007. DOI:10.1101/pdb.top17
- Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006; 116: 1767-1775. DOI: 10.1172/JCI29027
- Zhao FQ, Keating AF. Expression and regulation of glucose transporters in the bovine mammary gland. J Dairy Sci. 2007; 90: E76-E86. DOI: 10.3168/jds.2006-470
- Scheepers A, Joost HG, Schurmann A. The glucose transporter families SGLT and GLUT: Molecular basis of normal and aberrant function. J Parenter Enter Nutr. 2004; 28: 364-371. DOI: 10.1177/0148607104028005364
- Burant CF, Sivitz WI, Fukumoto H, Kayano T, Nagamatsu S, Seino S, et al. Mammalian glucose transporters: Structure and molecular regulation. In: Proceedings of the 1990 Laurentian Hormone Conference. 1991. P. 349-388.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4: 406-425. DOI: 10.1093/oxfordjournals.molbev.a040454
- Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981; 17: 368-376. https://tinyurl.com/yyust9pm
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33: 1870-1874. DOI: 10.1093/molbev/msw054
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution [N Y]. 1985; 39: 783-791. DOI: 10.1111/j.1558-5646.1985.tb00420.x
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018; 35: 1547-1549. DOI: 10.1093/molbev/msy096
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford university press; 2000.
- Das JK, Pal Choudhury P. Chemical property based sequence characterization of PpcA and its homolog proteins PpcB-E: A mathematical approach. PLoS One. 2017; 12: e0175031. DOI: 10.1371/journal.pone.0175031
- Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30: 1312-1313. DOI: 10.1093/bioinformatics/btu033
- Olsen GJ, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: A tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Bioinformatics. 1994; 10: 41-48. https://tinyurl.com/y5k3p87c
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004; 5: 389-396. DOI: 10.1038/nrg1327
- Liu M, Wu S, Walch H, Grigoriev A. Exon-domain correlation and its corollaries. Bioinformatics. 2005; 21: 3213-3216. DOI: 10.1093/bioinformatics/bti509


Sign up for Article Alerts