Proteomic Biomarkers as Indicator of Aquatic Pollution in Chrysichthys nigrodigitatus from Two Polluted Lagoons?
Bassey O.B1*, Chukwu L O1, Srivastava S2 and Reddy J2
1Department of Marine Sciences, Ecotoxicology laboratory, Faculty of Science, University of Lagos, Nigeria
2Proteomics laboratory, Faculty of Bioscience and Bioengineering, Indian Institute of Technology Bombay (IITB), Powai 400076 Mumbai, Maharashtra India
*Address for Correspondence: Bassey O.B, Department of Marine Sciences, Ecotoxicology laboratory, Faculty of Science, University of Lagos, Nigeria, Tel: +234-806-701-9883; E-mail: basseyob@gmail.com
Submitted: 23 July 2019; Approved: 06 August 2019; Published: 13 August 2019
Citation this article: Bassey OB, Chukwu LO, Srivastava S, Reddy J. Proteomic Biomarkers as Indicator of Aquatic Pollution in Chrysichthys nigrodigitatus from Two Polluted Lagoons. Int J Proteom Bioinform. 2019;4(1): 015-026.
Copyright: © 2019 Bassey OB, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: Chrysichthys nigrodigitatus; Proteomics; 2-D electrophoresis; MALDI-TOF-MS, iTRAQ, Lagoons
Download Fulltext PDF
Studying the expression of proteins in Chrysichthys nigrodigitatus muscle, is essential to understand the biological, physiological and ecological aspects that may be of advantage in ecotoxicology as a tool for biomonitoring the effects of environmental pollution, as well as food safety. This study was aimed to accomplish a systematic characterization of the muscle proteome as well as to identify a putative set of protein biomarkers in C. nigrodigitatus to environmental pollution in Ologe and Badagry lagoons. Fifteen fish samples were used as representative of the population for proteomics analysis. 116 proteins was expressed with 70 up-regulated, 25 down-regulated from Ologe Lagoon and 30 up-regulated, 17 down-regulated proteins from Badagry Lagoon expressed in the muscle of C. nigrodigitatus in response to environmental stressors using iTRAQ, while 8 protein spots from 2-D gels, representing 8 proteins with 2 up-regulated and 6 down-regulated, have been identified using MALDI TOF/TOF MS. Pearson correlation revealed significant correlation (p < 0.05) between environmental variables and protein markers. The investigation revealed that the expressed proteins in the muscle of C. nigrodigitatus served as a prognostic tool to assess the fish health and pollution status, which was observed that the fish were physiologically perturbed by environmental stressors in Ologe and Badagry lagoons.
Introduction
Aquatic ecosystems have been faced with several threats from complex mixture of contaminants from industries, anthropogenic perturbations and other stressors in recent years. And for several decades, the presence of these environmental contaminants in aquatic biotic and abiotic samples has been measured using chemical analytical techniques.
Environmental stress, as well as a variety of physical conditions, may sometimes induce the synthesis of certain proteins in fish. Some of these proteins are believed to play a role in protecting the cell from the damage, which may result from environmental perturbations, while others are involved in the regulation of various genes. The best-known representatives of this group are the stress proteins (also known as Heat Shock Proteins, (HSPs) and Metallothioneins (MTs).
Proteomics has also been applied to specific aspects of seafood research and technology. Proteomics applications related with seafood production and safety has been classified in three main items: (i) food safety studies, (ii) authentication and taxonomic applications, and (iii) nutritional aspects [1]. Proteins can be related to pollution initiation, safety, quality and nutrition topics, constituting in this sense important biomarkers for the detection of pollution, food safety, and nutritional value [2], as well as monitoring nutritional differences between wild and farmed species as applied in nutriproteomics [3].
Muscle plays a central role in whole-body protein metabolism by serving as the principal reservoir for amino acids to maintain protein synthesis in vital tissues and organs [4]. Skeletal muscle fibers represent one of the most abundant cell types in vertebrates [5] and contractile fibers of skeletal muscle tissues provide coordinated excitation-contraction-relaxation cycles for voluntary movements and postural control [6]. Besides playing a central physiological role in heat homeostasis, it usually presenting itself as a crucial metabolic tissue that integrates various biochemical pathways [7]. Therefore, muscle proteomics aims at global identification, cataloging and biochemical characterization of the entire protein complement of the voluntary contractile tissue as well as have the potential to identify novel proteins which could serve as biomarkers for many aspects including fish physiology and growth, flesh quality, food safety and aquatic environmental monitoring [8,9].
The most common methods for protein identification are Matrix-Assisted Laser Desorption Ionization- Time of Flight (MALDI-TOF) coupled with 2 Dimensional gel Electrophoresis (2DE) and Electrospray Ionization Tandem Mass Spectrometry (ESI-MS/MS). MALDI-TOF provides high sequenced coverage in proteins from sequenced genomes, while ESI-MS/MS is the method of choice in the case of genomes with no fully sequencing information available. The growth of mass spectrometry facilities, powerful bioinformatics tools, and availability of genomic information in research centres and industries can make possible that high throughput proteomics will feature in aquaculture and fisheries in the near future. The high-throughput proteomic approaches such as isobaric Tagging for Relative and Absolute Quantitation (iTRAQ), has been successfully used to study the responses by fish to aquatic pollutants, including androgen receptor agonists/antagonists [10]. This high-through proteomics has been successfully applied to identify altered protein expression in the muscle of rainbow trout Oncorhynchus mykiss during spawning and was associated with biochemical processes involved in muscle deterioration.
Proteomic in Fish
It is increasingly important to profile proteins in order to understand biological processes in a post genomic era as the dynamics of proteins between cells at different times and under different environmental conditions provide an actual biological phenotype. Proteomics studies are built upon the foundations of genomics, in such way that a lack of genomic information on a particular species can substantially limit the success in protein identification [11]. As such, functional genomics and proteomic technologies have allowed simultaneous biological study of thousands of genes or proteins. In particular, the presence of post translational modifications in proteins further highlights the importance of proteomic analysis which is not replaceable by other genomic approaches [12].
Techniques and Approaches in Identification of Potential Protein Markers
Different molecular approaches [biochemical assays, Enzyme Linked Immuno-Sorbent Assays (ELISA), spectrophotometric, fluorometric measurement, differential pulsed polarography, liquid chromatography, atomic absorption spectrometry]. However, methodological approaches able to detect subtle changes in the expression of individual proteins and amino acid sequence modifications, constitute an excellent tool [13,14], which can be explanatory of a biological situation. Recent report shows exponential developments concerning proteome analytical techniques and sample preparation [15]. Proteomics is essentially based on classical “Analytical Chemistry” strategies: separation by electrophoresis techniques, identification and quantitative analysis through mass spectrometry.
Proteomics as Environmental Marker of Pollution
Proteomics applied to assess the biological effects of pollutants in marine organisms is beginning to reveal some of the systemic changes that occur on the cellular level. First, even a qualitative description of cellular changes through PES shows that the systemic changes occurring during exposure are pollutant-specific. Second, proteins common to many pollutant-stress responses include oxidative stress proteins, cytoskeletal proteins, chaperones, proteases, and proteins involved in the detoxification of xenobiotics as well as β-oxidation. Together, these changes suggest that the production of ROS leads to the denaturation of proteins as well as wide-ranging modifications of cytoskeletal elements. Finally, PTMs present a novel frontier to assess the biological effects of pollutants. Among the PTMs that are likely to be the most important are those that increase with oxidative stress, e.g. Carbonylation and glutathionylation, and that are indicators of protein denaturation, e.g. ubiquitination.
Fisheries have received a great deal of concern as a potential source of heavy metal contamination to humans because of its wide consumption of fisheries [16,17]. In West Africa, the Bagrid catfish (Chrysichthys nigrodigitatus) are commercially important fish species that are widely consumed and continue to attract high patronage. Chrysichthys nigrodigitatus on the other hand, is a demersal omnivore inhabiting the bottom of shallow waters, rivers and lagoons [18].
The Ologe and Badagry Lagoons, a home for a wide variety of fishery resources have been found to be subjected to many anthropogenic sources of contamination by virtue of their socio-economic importance and proximity to urban centres, heavy-industrial and use for several agricultural activities. Previous studies [19-23] revealed varying levels of contaminants in these ecosystems including heavy metals, pesticides and polycyclic aromatic hydrocarbons. These compounds can be absorbed and accumulated in the edible parts of lagoon biota, thus entering the human food chain and posing a public health problem. However, there is paucity of information on the proteome compositions of most edible aquatic biota from Ologe and Badagry Lagoons. Therefore, proteomic studies would be quite useful as additional information during environmental contamination studies in these ecosystems.
The aim of this study was to evaluate the potential of proteomics as a tool for biomarker discovery, providing characterization of the proteome sensitive pollution markers in the muscle of C. nigrodigitatus captured from the wild. Thus, the application of proteomic hypothesis-independent approach was to discover the protein expression patterns underlying the effects of environmental stressors in form of admixtures of chemical pollutants from several anthropogenic activities in an attempt of identifying a putative set of protein biomarkers.
Materials and Methods
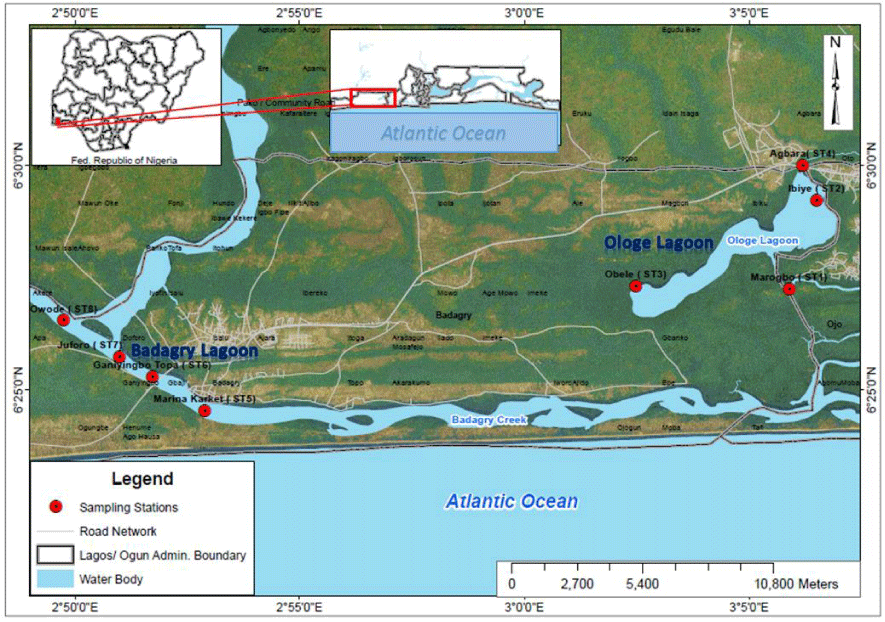
Ologe and Badagry Lagoons are situated within the Lagos lagoon complex in Lagos State of Southwestern region of Nigeria (Figure 1). Ologe Lagoon is a freshwater body which opens into the Atlantic Ocean via the Badagry creeks and the Lagos harbour. These Lagoons meet several socio-economic needs such as aquaculture, fishing, sand dredging and drainage across various towns and villages bordering it [19]. By virtue of their position, (with several industries and other anthropogenic activities around it), have been impacted by partially treated and untreated industrial effluents.
Experimental set-up
Fish (Chrysichthys nigrodigitatus) samples were collected using a trap basket slightly above the bottom of the lagoon, where the water was shallow at depth of 3.5 m from the eight (8) established stations in Ologe and Badagry Lagoons between October 2013- April 2015.
Water quality and heavy metal analyses
Water quality parameters were characterized following APHA standard protocols [24]. Two (2) g of the dried sediment was taken in a digestion beaker and 10 mL of mixture of HCl and HNO3 in the ratio 3:1 was added and then digested at 95°C for 2 h. The metal concentrations of both the digested water and sediment samples were quantified using GBC (Savant AA Sigma) flame Atomic Absorption Spectrometer (AAS). All chemical regents used herein were of analytical reagent grade (Merck, United State). The glassware was pre-cleaned with nitric acid and rinsed with double distilled water.
Preparation of protein extracts and quantification
One hundred (100) mg of muscle tissue were collected from Chrysichthys nigrodigitatus and minced using a clean scalpel. The tissue was homogenized with 1 mL of PBS and spinned at 3,000 rpm for 10 min at 40C. The supernatants were removed, and then 1 ml of whole cell lysis buffer was added to the 100 mg of homogenized tissue. The Whole Cell Lysis (WCL) is composed of 50 mM NaCl, 1M HEPES (pH 7.8), 5 mg/ml leupeptin, 10 mg/ml Pepstatin, 250 mM DDT, Tris, 100 mM PMSF, 40 mM Na3VO4 and distilled water. Sonication for 4x8 at output level 5, Spinned at 13,000 rpm for 10 min at 40C, the supernatant was removed, saved in another tube and centrifuged. The mixture was incubated in ice for 30 additional minutes, and then divided in 100 μL aliquots that were stored at -80°C until needed. The quantification results and the quality of the protein extracts was performed by the modified Bradford method, measuring the samples extracts in duplicates and using Bovine Serum Albumin (BSA) as protein standard. The results of the quantification were checked by running 25 μg of each protein extract on 12.5% vertical SDS-PAGE gels according to Laemmli’s protocol [25].
For 2D-PAGE, protein extracts were diluted in rehydration buffer with composition 60 mM urea, 2 M thiourea, 2% CHAPS, 1% DTT, 0.5% IPG Buffer pH 3-10. Resuspended proteins, approximately 300 μg, were absorbed overnight into 13 cm Immobilized Dry strip pH 3-10 (GE Healthcare) and focused at 50 μA per strip according to the manufacturer’s protocol, for a total of 78,300 Vh in an IPGphor isoelectrofocusing unit (GE Healthcare). After focusing, strips were equilibrated with equilibration buffer (6 M Urea, 75 mM Tris HCl, pH 8.8, 29.3% (v/v), 30% glycerol, 2% (w/v) SDS and 0.002% (w/v) Bromophenol blue containing 10 mg/ml supplemented with 1% DTT for 15 min, and then with 2.5% iodoacetamide for 15 min on roller mixer.
The Second Dimension (SDS-PAGE) [23] run was performed using 12.5% resolving gels with 6% (w/v) stacking gel on a Protean-2 cell (Bio-Rad). The gels were stained with Coomassie Brilliant Blue (CBB), and images were acquired by Image Scanner LabScan 6.0 (GE Healthcare Biosciences) Proteomic Lab, IITB, India. The quantification of protein was estimated using the Bradford Method and standardized with BSA (Bovine Serum Albumin). The quantification of protein extracts observed low concentrations across individual replicate samples in each seasons.
1-D SDS-PAGE gels were used to examine the range of protein MW and to assess the presence of interfering substances in the muscle protein extracts. The standard of protein determined from absorbance at 595nm wavelength plotted against Bovine Serum Album (BSA) concentrations.
In-Gel Digestion
In-gel digestion using trypsin was performed according to Shevchenko, et al. [26]. The polyacrylamide gel was washed thoroughly with 100 mM NH4HCO3. The protein bands were then excised from the gel.
Matrix Laser Desorption Ionization Time of Flight/ Time Of Flight Mass Spectrometry (MALDI-TOF/TOF-MS)
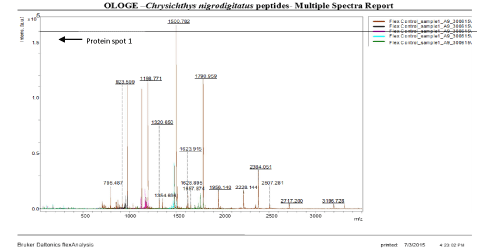
A total of 8 gel spots were excised from the 2D gels, for MALDI-TOF/TOF-MS analysis; gel spots were picked up from one freshly run gel. The gel spots were destained and digested overnight with trypsin. The resulting peptides were extracted following standard techniques [27]. Peptide fragment mass spectra were acquired in data dependent Bruker Daltonic Flex analysis with a scan range of 500–3500 m/z; three averages and up to three precursor ions were selected from the MS scan (100–3500 m/z). Protein mass score is -10*Log (P), where P is the probability that the observed match is a random event. Protein scores greater than 46 were significant (p < 0.05).
Sequence database search
The MS/MS data were subjected to Mascot protein database search engine (www.matrixscience.com) [28]. The search engine contains the calculated spectra for all peptides in the National Centre for Biotechnology Information (NCBI) non-redundant sequences database (ncbi.nlm.nih.gov) and UniProt (uniprot.org/).
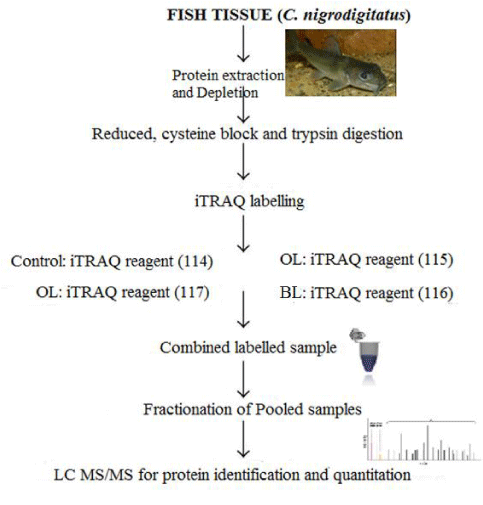
Isobaric tag relative and absolute quantitation (iTRAQ) labeling
Peptide fractionation was performed using a strong cation exchange column as described by Zhang, et al. [29]. Peptides of the C. nigrodigitatus from Ologe and Badagry lagoon were labeled with the 114, 115, 116, and 117 iTRAQ reporters, respectively. The fractioned peptide samples were dried, while the columns prepared were activated with 1 ml of Acetonitrile (ACN), 0.1% Formic acid and a combined 50% (1 ml ACN) and 50% (0.1% FA) and then washed with 0.1% FA. iTRAQ was performed using Acquisition Mode AutoMS2 with MS Range: 300-3000 m/z MS Q-TOF with ModelG6550A (Agilent Technologies). The 4-plex iTRAQ is a reagent used to label all protein samples in the muscle of C. nigrodigitatus from Ologe and Badagry Lagoons. A total of 12 representative protein samples for the 4-plex iTRAQ analysis of C. nigrodigitatus muscles from Ologe and Badagry Lagoon was quantified, pooled together, fractionated and analyzed to identify the peptide sequence in the tandem mass spectrometry.
Result and Discussion
Water quality characteristics
The mean variations of physico-chemical properties of water quality of the lagoons are presented in table 1. The water quality parameters indicates significant differences (p < 0.05) across sampling period in each lagoons. Furthermore, T-Test revealed significant differences (p < 0.05) in pH, Salinity, Dissolved Oxygen (DO) and Biochemical Oxygen Demand (BOD) between the two Lagoons across the sampling period. The DO and BOD levels in the water fell below permissible limit of FMEnv (2003).
Table 2 showed the mean levels of heavy metals in sediment at Ologe and Badagry lagoons. ANOVA revealed that all metals in the sediment, except Ni showed significant differences (p < 0.05) in both lagoons. The T-test also indicated that there was a significant difference (p < 0.05) in the metals of both lagoons. Furthermore, Pearson correlation coefficient (Table 4) showed significant correlation between environmental variables and protein markers (over-expression: Up-regulated and suppressed: down-regulated proteins).
Table 4: Pearson correlation coefficient of physico-chemical properties of the water, heavy metals in sediment and up and down regulated proteins in C. nigrodigitatus from Ologe and Badagry lagoons.
Protein identification by Matrix-Assisted Laser Desorption/ Ionization Time of Flight/ Time of Flight Mass Spectrometry (MALDI-TOF/TOF-MS)
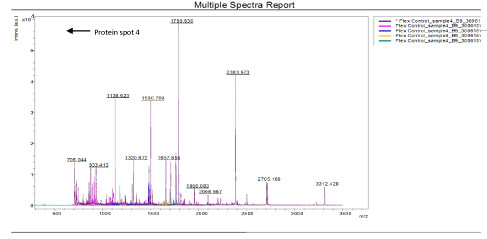
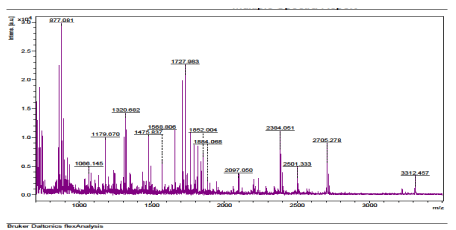
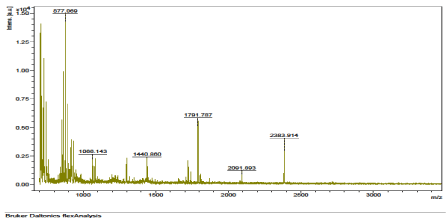
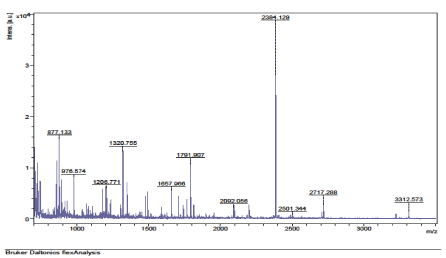
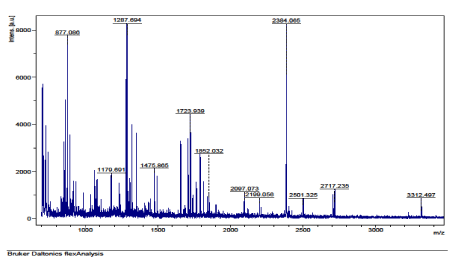
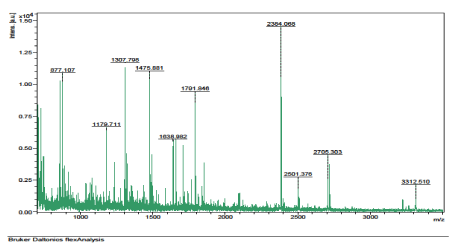
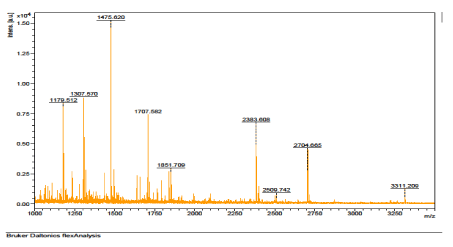
The results of the analysis on Matrix-Assisted Laser Desorption/ Ionization Time of flight/ Time of flight mass spectrometry (MALDI-TOF/TOF-MS) carried out for the detection, identification and characterization of protein markers. From the 2D gel run for spot picking for MALDI-TOF/TOF-MS, 8 spots were visualized on the CBB-stained gels of the muscle protein extract of C. nigrodigitatus (plate 1). The identified 8 protein spots were subjected to Mascot search engine database for fish species. Of the eight, 2 protein spots (1 and 4) were involved in cell structure. The MS/MS data (peptide spectra) subjected to Mascot protein database search engine (www.matrixscience.com) also identified proteins with mass score greater than 46 which indicates that there was a significant difference (p < 0.05) and that they were over-expressed (up-regulated). On the other hand, protein mass score below 46 indicated no significant difference (p > 0.05) and were down-regulated. At Badagry lagoon, the spectra and gel map had protein mass scores below the required threshold level of 46. Spectra are displayed in Figured 5, 6, 7, 8, 9 and 10 had no significant protein mass scores.
Further analysis indicated that, 2 proteins spots represented by cytoskeletal proteins, Alpha Actin and Actin cytoplasmic 1 (spots 1 and 4) were significantly different (p < 0.05) indicating over-expression in the muscle protein (Plate 1). On the other hand, Fructose-bisphosphate aldolase A (spot 3), Spot 2 and 8: Metallothionein, spot 5: Parvalbumin beta 1, Spot 6: Hemoglobin subunit beta-1, and Spot 7: Prolactin-1 showed no significant difference (p > 0.05) in muscle of C. nigrodigitatus indicating that the proteins from spot 2, 3, 5, 6, 7 and 8 were down-regulated in the muscle (Figure 10-15). The peptide spectra of the cytoskeletal proteins of spot 1 and 4 that are over-expressed are presented in figure 3 & 4.
Protein identification by isobaric Tag for Relative and Absolute Quantitation (iTRAQ)
The measurement of protein responses to contaminants in muscle tissue of C. nigrodigitatus using iTRAQ detected 116 peptide sequences. The peptide sequences after subjection to tandem Mass Spectrometry/ Mass Spectrometry in a pool across sample stations and seasons revealed the identity of potential protein markers in muscles of C. nigrodigitatus from Ologe and Badagry lagoons. Further evaluation of the 116 identified peptide sequences subjected to the National Centre for Biotechnology Information (NCBI) database provided little or no information on the protein sequence for Chrysichthys nigrodigitatus in the database. However, on evaluation using UniProt database search engine, the gene identity of the protein markers was revealed based on their relationship with the molecular weight and gene expressions from other related catfishes (Table 3).
A total of 100 (86.20%) proteins were observed to be over-expressed (up-regulated) in the muscle of C. nigrodigitatus as a consequent of thermal stress and hypoxic conditions influenced by various environmental factors representing 60.34% and 25.86% in the muscle of C. nigrodigitatus from Ologe and Badagry lagoons respectively.
The results also indicated that, of the 116 identified proteins in the muscle of C. nigrodigitatus 70 proteins were up-regulated and 25 proteins down-regulated in Ologe lagoon, while 30 proteins were over-expressed and 17 proteins were down-regulated in Badagry lagoon.
The observed up-regulated proteins were identified as; Heat shock protein- Hsp 70 and 90, Lactate Dehydrogenase A-chain (LDH), Creatine Kinase (CK), cytochrome c oxidase, regucalcin. Similarly, total of 47 (40.51%) proteins were observed to be depressed (down-regulated) in the muscle of C. nigrodigitatus representing 53.19% and 36.17% in the muscle of C. nigrodigitatus from Ologe and Badagry lagoons respectively. The observed down-regulated proteins were identified as cytochrome b, Triosephosphate, alpha-β crystalline, and Tropomyosin isoforms as presented in table 3.
Functional groups and biological pathways of the identified proteins in the muscle of C. nigrodigitatus
The identified proteins were assigned to several functional groups with respect to their involvement with cell structures, signal transduction, enzyme regulation, and oxidative stress as follows: alpha actinin, alanine aminotransferase, heat shock protein HSP 90-alpha 1, transmembrane, and β-tubulin, among others as presented in figure 11.
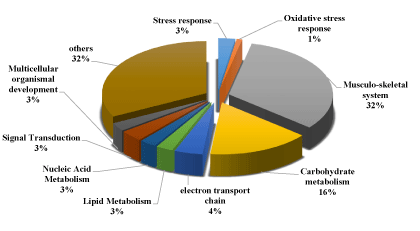
The identified proteins were divided into ten categories, viz. cytoskeletal proteins, carbohydrate metabolism, nucleotide metabolism, defense response to oxidative stress, response to stimulus (Heat shock protein), multicellular organismal development, signal transduction, electron transport chain, lipid metabolism, and others.
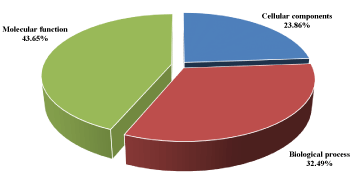
The results of the gene ontology analyses of identified proteins in the muscle tissue of C. nigrodigitatus also showed that 32.49% were involved in biological processes, 43.65% in molecular functions and 23.86% in cellular components. Of the 32.49% proteins involved in biological processes, 32% were associated with musculo-skeletal system, 16% accounting for carbohydrate metabolism, responses to stimuli accounted for 3% and 1% of the proteins related to oxidative stress response as presented in figure 12.
Discussion
The proteomics biomarker of environmental pollution in Chrysichthys nigrodigitatus was evaluated at Ologe and Badagry Lagoon between Oct., 2013 and Apr., 2015. Proteomics, the global analysis of protein synthesis, studies the protein expression patterns in response to environmental change. Two-dimensional protein gels, combined with peptide mass mapping by MALDI TOF MS for protein identification, as well as iTRAQ was used for determining differential protein synthesis in biological systems.
The results of the 2-D gel electrophoresis and MALDI-TOF/TOF MS carried out to investigate the proteome composition and identify protein expression patterns revealed a total of 8 individual spots on the basis of their Peptide Mass Fingerprints (PMF). This represented 2 proteins peptide identified as α-Actin and Actin cytoplasmic 1. These protein markers was also observed to be over-expressed, indicating the attempt of maintaining cell shape and functionality in cases of hypoxic and heat shock stress.
The proteomic iTRAQ analysis, a 4-plex labeled sample from Ologe and Badagry Lagoons identified and quantified 116 protein markers. A total of 70 up-regulated and 25 down-regulated proteins in the muscles of C. nigrodigitatus from Ologe Lagoon and 30 up-regulated and 17 down-regulated proteins in the muscle of C. nigrodigitatus from Badagry Lagoon. This is an indication of changes in the protein expressions (depressed and over-expressed) in the muscle of C. nigrodigitatus induced by oxidative stress, heat shock and other variables of environmental stress in an environmentally polluted Lagoon. Adaptation with the environmental stress leads to elevated expression of heat shock genes that produce heat shock proteins, which in turn interact with stress-denatured proteins to maintain or restore their native structures and prevent aggregation and degradation [30,31].
The α-tubulin proteins are linked to heavy metal tolerance [32,33], which was exemplified in this study where α-tubulin 1 was significantly enhanced in the muscle with an increase protein fold (over-expressed) in response to the heavy metal in the lagoons. Keratins are important intermediate filament proteins, and their primary function is to protect the cells from stress damage that may result in cell death [34,35]. The up-regulation of keratins in the muscle of C. nigrodigitatus was expected to fight against oxidative stress induction from both lagoons.
Actins are highly conserved proteins that are involved in various important cellular processes including cell motility, cell signaling, and the establishment and maintenance of cell junctions and cell shape. The change of actin isoforms (α- and β-Actin) in abundance might have been related to induce oxidative stress in Ologe and Badagry lagoons, since Actins can be a direct target for oxidative modification [36]. Actin is a cytoskeletal protein that is ubiquitously expressed in many eukaryotic cells and functions as maintenance of the cytoskeleton, cell motility and muscle contraction. The changes in the level of cytoskeletal and structural proteins expression can be very often attributed to the attempt of maintaining cell shape and functionality in cases of hypoxic [37,38] associated with low dissolved oxygen and slightly acidic pH level of the water quality, which is indication of an environmentally stressed Ologe lagoon, as compared to Badagry Lagoon indicating no significant difference (p > 0.05) of the protein markers in the muscle of C. nigrodigitatus.
The Metallothionein (MT) was identified to be down-regulated in the muscle of C. nigrodigitatus. MT is a ubiquitous, low molecular weight, cysteine-rich (>30%) protein that avidly binds various transition elements. Metallothionein is an excellent environmental biomarker. Stress caused by both bio-active and non-bioactive metals induces a variety of complex changes in fish physiology and the possible consequent physiological alterations as observed in C. nigrodigitatus from Ologe and Badagry lagoons.
Another identified protein was Parvalbumin beta 1, involved in relaxation after contraction of the muscles. It binds two calcium ions and also involves in calcium signaling. The Parvalbumin was observed to be down-regulated in the muscle which disrupts the involvement at the calcium binding site, and relaxation of the muscles which might be induced by environmental stressors. Prolactin was also identified to be down-regulated. Prolactin 1 is a protein hormone of the anterior pituitary gland.
HSP-90 belongs to the Heat Shock Protein Family (HSPs), which are well known to protect the structure and function of cells from stress (e.g. metal attack, climate change, thermal stress and hypoxic conditions generated from other environmental factors) and play an important role in maintaining cellular homeostasis [39]. The deletion of HSP-90 is lethal for eukaryotic cells [40], and in this study, the over-expressed of HSP-90 suggested that the muscle of C. nigrodigitatus inhibited the stressed condition by enhancing the Hsp in the Ologe lagoon, while at Badagry lagoon the heat shock protein beta 1 was observed to be suppressed.
Enolase is a cytosolic enzyme involved in carbohydrate metabolism, cell differentiation, and normal growth, and a decline of enolase activity results in abnormal growth and reduced metabolism in the muscle. The enolase 1 alpha, partial and beta-enolase were up-regulated in the muscle which indicates an active metabolism with normal growth of the fish species in the lagoons. Creatine kinase is an essential enzyme for energy buffering to maintain cellular energy homeostasis. Its activity is inhibited by metals [41]. The increased (up-regulation) levels of enolase 1 (alpha) and creatine kinase in the muscles of C. nigrodigitatus are an adaptive feedback to inhibitory activity in the muscle under oxidative stress. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. The over expression of Cytochrome C Oxidase (COX2) in muscle proteome of C. nigrodigitatus, is an indication of adaptation to hypoxic condition induced by admixture of environmental pollutants in the dry season at Ologe lagoon.
Regucalcin was identified to be over-expressed in the muscle of C. nigrodigitatus in both lagoons. Regucalcin has been found to play a multifunctional role in different tissues, and is primarily involved in the maintenance of intracellular Ca2+ homeostasis (calcium ion binding and enzyme regulator activity). Over-expression of regucalcin are usually found to enhance glucose utilization and lipid production in fish when exposed to severe oxidative stress observed in Ologe and Badagry Lagoons respectively. Cytochrome b protein are component of the ubiquinol - cytochrome c reductase complex, which functions as respiratory electron transport chain that generates an electrochemical potential coupled to ATP synthesis. The over-expression of the cytochrome b (cyt b) indicates a significant decline of dissolved oxygen, rendering the cyt b to increase its electron transport chain for respiratory functions as an adaptive mechanism in the muscle of C. nigrodigitatus at the dry season in Ologe Lagoon.
Another identified protein that was over-expressed is Lactate Dehydrogenase A-Chain (LDH), is a key enzyme in the control of energy metabolism, catalyzing the interconversion of pyruvate to lactate and regulating the levels of these metabolites in accordance with oxygen availability. Under anaerobic conditions, the isoform LDH-A4 (isozyme A4) preferentially converts pyruvate to lactate. This isoform is found predominantly in poorly vascularized tissues with low partial Pressure of Oxygen (pO2), such as skeletal muscle in both lagoons and seasons.
Triosephosphate Isomerase (TPI) was observed to be down-regulated in the muscle of C. nigrodigitatus. TPI is a glycolytic enzyme that catalyzes the conversion between glyceraldehyde-3-phosphate and dihydroxyacetone phosphate; it is essential for efficient energy production by being involved in several metabolic pathways. The significant changes of TPI expression might alter catalytic process of energy production induced by pollutants in Ologe Lagoon. Another identified down-regulated protein was α-B crystalline, which is responsible for metal ion binding, structural constituents for the eye lens. The less abundant corneal proteins might be unable to contribute structurally to its transparency and optical properties of C. nigrodigitatus at the Badagry Lagoon.
Phosphoglucomutase-1 was to be down-regulated in the muscle of C. nigrodigitatus in Ologe lagoon. Phosphoglucomutase-1 is a key enzyme in the metabolism of glycogen and protein glycosylation. It is responsible for the reversible inter conversion of glucose 1-phosphate to glucose 6-phosphate, both of which are key intermediates in the synthesis and breakdown of glycogen and galactose metabolism. It is also important for the formation of UDP-glucose which is an essential intermediary metabolite in protein glycosylation. Inhibition of phosphoglucomutase has drastic effects on carbohydrate metabolism which reduces the steady-state levels of UDP-glucose, resulting in a defect of glycogen and trehalose biosynthesis, while galactose metabolism is inhibited, leading to galactosemia, accumulation of galactose 1-phosphate and Glucose 1-phosphate i.e., poor glycogen turnover.
Tropomyosin alpha-1 was observed to be up-regulated in both lagoons, while Tropomyosin alpha-3 chain was observed to be down-regulated as well in both lagoons and Tropomyosin isoforms have been reported both in condition of thermal stress [37] and hypoxia [38].
Conclusion
This study identified protein markers that are over-expressed (Up-regulated) and depressed (Down-regulated) in the muscle of C. nigrodigitatus which serves as prognostic tools (MALDI-TOF/TOF and iTRAQ approach) to assess the pollution status of Ologe and Badagry lagoons, and useful for biotechnological interventions in fish health and disease management; besides adding to the existing knowledge base on comparative muscle proteomics on C. nigrodigitatus of a tropical ecosystem.
Acknowledgement
The author appreciate the mentorship and support of Prof. Chukwu, L. O. of Department of Marine Sciences, University of Lagos, Prof. Sanjeeva S. and all Ph.D. colleagues of department of Bioengineering and Bioinformatics, Proteomics unit, Indian Institute of Technology Bombay (IITB), Powai Mumbai India.
- Pineiro C, Canas B, Carrera M. The role of proteomics in the study of the influence of climate change on seafood products. Food Research International. 2010; 43: 1791-1802. http://bit.ly/2KJv0S7
- Thomas T, Egan S, Burg D, Ng C, Ting L, Cavicchioli R. Integration of genomics and proteomics into marine microbial ecology. Marine Ecology Progress Series. 2007; 332: 291-299. http://bit.ly/2ZWlMbA
- Reddish JM, St Pierre N, Nichols A, Green Church K, Wick M. Proteomic analysis of proteins associated with body mass and length in yellow perch, Perca flavescens. Proteomics. 2008; 8: 2333-2343
- Mohanty BP, Banerjee S, Bhattacharjee S, Mitra T, Purohit GK, Sharma AP, et al. Muscle Proteomics of the Indian Major Carp Catla (Catla catla, Hamilton). J Proteomics Bioinform. 2013; 6: 252-263. http://bit.ly/2KvmE1K
- Ohlendieck K. Proteomic profiling of fast-to-slow muscle transitions during aging. Front Physiol. 2011;2: 105. http://bit.ly/2Ktf5Zu
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80: 853-924. http://bit.ly/33lQ9KK
- Abdul Ghani MA, DeFronzo RA. Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol. 2010; 47: 62- 79. http://bit.ly/2yUWsXG
- Addis MF, Cappuccinelli R, Tedde V, Pagnozzi D, Porcu MC. Proteomic analysis of muscle tissue from gilthead sea bream (Sparus aurata, L.) farmed in offshore floating cages. Aquaculture. 2010; 309: 245-252.
- Barik SK, Banerjee S, Bhattacharjee S, Das Gupta SK, Mohanty S, Mohanty BP. Proteomic analysis of sarcoplasmic peptides of two related fish species for food authentication. Appl Biochem Biotechnol. 2013; 171: 1011-1021. http://bit.ly/2YIkvZ7
- Martyniuk CJ, Alvarez S, McClung S, Villeneuve DL, Ankley GT, Denslow ND. Quantitative proteomic profiles of androgen receptor signaling in the liver of fathead minnows (Pimephales promelas). J Proteome Res. 2009; 8: 2186-2200. http://bit.ly/2yUWIpC
- Graham DR, Elliott ST, Van Eyk JE. Broad-based proteomic strategies: A practical guide to proteomics and functional screening. J Physiol. 2005; 563: 1-9. http://bit.ly/2KIxekO
- Stroncek DF, Burns C, Martin BM, Rossi L, Marincola FM, Panelli MC. Advancing cancer biotherapy with proteomics. J Immunother. 2005; 28: 183-192.
- Ahmed, FE. Utility of mass spectrometry for proteome analysis: Part I. Conceptual and experimental approaches. Expert Rev Proteomics. 2008; 5: 841-864. http://bit.ly/2YILVOO
- Nesatyy VJ, Suter MJ. Analysis of environmental stress response on the proteome level. Mass Spectrometry Reviews. 2008; 27: 556-574.
- Canas B, Pineiro C, Calvo E, Lopez Ferrer D, Gallardo JM. Trends in sample preparation for classical and second generation proteomics. J Chromatogr A. 2007; 1153: 235-258. http://bit.ly/2ZT44pG
- Donkor, K A, Bonzongo J J, Nartey K V, Adotey K D. Heavy metals in sediments of the gold mining impacted Pra River Basin, Ghana, West Africa. Soil Sediment Contam. 2005; 14: 6, 479-503.
- Gbogbo F, Otoo DS. The concentrations of five heavy metals in components of an economically important urban coastal wetland in Ghana: public health and phytoremediation implications. Environ Monit Assess. 2015; 187: 655. http://bit.ly/2KEHL0u
- Offem BO, Akegbejo Samsons Y, Omoniyi IT. Diet, size and reproductive biology of the silver catfish, Chrysichthys nigrodigitatus (Siluriformes: Bagridae) in the Cross River, Nigeria. Rev Biol Trop. 2008; 56: 1785-1799. http://bit.ly/2H0T8Pm
- Clarke EO, Anetekhai MA, Akin Oriola GA, Onanuga A I S, Olarinmoye, OM, Adeboyejo OA, Agboola I. The Diatom (Bacillariophyta) diversity of an open access lagoon in Lagos, Nigeria. J Res Rev Sci. 2004; 3: 70-77.
- Albaret J, R Lae. Impact of fishing on fish assemblages in tropical lagoons: The example of the Ebrie Lagoon, West Africa. Aquat Living Resour. 2003; 16: 1-9
- Anetekhai, MA, GA Akin Oriola, OJ Aderinola, SL Akintola. Trace metal concentration in Macrobrachium vollenhovenii from Ologe Lagoon, Lagos, Nigeria. J Afrotropical Zool. 2007; 3: 25-29.
- Agboola JI, MA Anetekhai, AAB Denloye. Aspects of the ecology and fishes of Badagry creek (Nigeria). J Fish Aquat Sci. 2008; 3: 184-194.
- Kumolu Johnson CA, PE Ndimele, SL Akintola, CC. Jibuike. Copper, zinc and iron concentrations in water, sediment and Cynothrissa mento (Regan 1917) from Ologe Lagoon, Lagos, Nigeria: A preliminary survey. Afr J Aquat Sci. 2010; 35: 87-94. http://bit.ly/33lP8lU
- APHA (2005) Standard methods for the examination of water and waste water (21st ed.),Washington, D.C: American Public Health Association.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680-685. http://bit.ly/2YWWk4t
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of protein from silver-stained polyacrylamide gels. Anal. Chem. 1996; 68: 850-858. http://bit.ly/2Z0wzjZ
- Bringans S, Eriksen S, Kendrick T, Gopalakrishnakone P, Livk A, Lock R, et al. Proteomic analysis of the venom of Heterometrus longimanus(Asian black scorpion). Proteomics. 2008; 8: 1081-1096. http://bit.ly/31E8eCh
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999; 20: 3551-3567. http://bit.ly/2ZZ5GhI
- Zhang H, Zhao C, Li X, Zhu Y, Gan C S, Wang Y, Ravasi T, Qian P Y, Wong SC, Sze S K. Induced tolerance using iTRAQ-based quantitative proteomic approach. Proteomics. 2010; 10: 2780-2789
- Ielmini SE, Piredda G, Mura S, Greppi GF. Protein biomarkers as indicator for water pollution in some lagoons of Sardinia. Transitional Waters Bulletin. 2014; 8: 32-52. http://bit.ly/2H2s9Tt
- Purohit GK, Mahanty A, Suar M. Investigating hsp gene expression in liver of Channa striatus under heat stress for understanding the upper thermal acclimation. Biomed Res Int. 2014; 14: 1-10. https://bit.ly/33t9pWG
- Mattingly KS, Beaty BJ, Mackie RS, McGaw M, Carlson JO, Rayms Keller A. Molecular cloning and characterization of metal responsive Chironomus tentans alpha-tubulin cDNA. Aquat Toxicol. 2001; 54: 249-260. https://bit.ly/33vjXob
- Mireji PO, Keating J, Hassanali A, Impoinvil DE, Mbogo CM, Muturi MN, et al. Expression of metallothionein and β-tubulin inheavymetal-tolerant Anopheles gambiae sensustricto (Diptera: Culicidae). Ecotox Environ Safe. 2010; 73: 46-50. https://bit.ly/2yTdky7
- Looi ML, Karsani SA, Rahman MA, Dali AZ, Ali SA, Ngah WZ, et al. Plasma proteome analysis of cervical intraepithe lial neoplasia and cervical squamous cell carcinoma. J Biosci. 2009; 34: 917-925. http://bit.ly/33wswPF
- Omary MB, Ku NO, Toivola DM. Keratins: guardians of the liver. Hepatology. 2002; 35: 251-257. https://bit.ly/2yTwbJj
- Lassing I, Schmitzberger F, Bjornstedt M, Holmgren A, Nordlund P, Schutt CE, et al. Molecular and structural basis for redox regulation of α-Actin. J Mol Biol. 2007; 370: 331-348. https://bit.ly/2KJkryw
- Tomanek L, Zuzov MJ. The proteomic response of the mussel congeners Mytilus galloprovincialis and M. trossolus to acute heat stress: implications for thermal tolerance limits and metabolic costs of thermal stress. J Exp Biol. 2010;213: 3559-3574. https://bit.ly/2ZWnNVc
- Letendre J, Dupont Rouzeyrol M, Hanquet AC, Durand F, Budzinski H, Chan Vaudry D, et al. Impact of toxicant exposure on the proteomic response to intertidal condition in Mytilus edulis. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6: 357-369. https://bit.ly/33v4lRA
- Sanders BM. Stress protein in aquatic organisms: an environmental perspective. Crit Rev Toxicol. 1993;23: 49-75. https://bit.ly/2Mdar3J
- Voss AL, Thomas T, Gruss P. Mice lacking hsp90 fail to develop a placental labyrinth. Development. 2000; 127: 1-11. https://bit.ly/2Kt5NfQ
- Glaser V, Leipnitz G, Straliotto MR, Oliveira J, dos Santos VV, Wann macher CM. et al. Oxidative stress-mediated inhibition of brain creatine kinase activity by methylmercury. Neurotoxicology. 2010; 31: 454-460. https://bit.ly/33tqZdq

Sign up for Article Alerts