Triton X-100 Vs. Triton X-114: Isolation of Outer Membrane Proteins from Leptospira Spp.?
Angel A. Noda1*, Osmel Fleitas1, Islay Rodriguez1, Jorge F. Beltran1, Rosabel Falcon1, Tatiana Almaguer1 and Samaha TH2
1Department of Bacteriology-Mycology, Institute of Tropical Medicine, “Pedro Kouri”, Havana, Cuba
2Regional Medical Research Centre, Indian Council of Medical Research, WHO Collaborating Centre for Leptospirosis, Port Blair, Andaman and Nicobar Islands, India
*Address for Correspondence: Angel A. Noda, Department of Bacteriology-Mycology, Institute of Tropical Medicine “Pedro Kouri”, Havana, Cuba, Tel: +537-255 3530; Fax: +537-204 6051; E-Mail: angelalberto@ipk.sld.cu
Submitted: 07 July 2017; Approved: 24 July 2017; Published: 27 July 2017
Citation this article: Noda AA, Fleitas O, Rodriguez I, Beltran JF, Falcon R, et al. Triton X-100 Vs. Triton X-114: Isolation of Outer Membrane Proteins from Leptospira Spp. Int J Vet Sci Technol. 2017;1(1): 007-012.
Copyright: © 2017 Noda AA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: Leptospira; Triton X-100; Triton X-114 ; Membrane Protein Isolation
Download Fulltext PDF
Leptospirosis, caused by Leptospira Spp., is a disease of worldwide significance affecting millions of people annually. The outer membrane of Leptospira facilitates direct interactions with the surroundings and contains important molecules involved in infection, transmission, survival, and adaptation to environmental conditions. The results showed that Triton X-100 can extract outer membrane protein without co extraction of inner membrane or cellular protein as evidenced by the absence of cell lysis. The comparison with existing Triton X-114 method showed that Triton X-100 is more selective in partitioning protein into the detergent phase. Our results establish a broadly applicable strategy for the preparation of outer membrane proteins of Leptospira, an essential step in the identification of potential virulence factors, diagnostic antigens and vaccine candidates.
Introduction
Leptospirosisis one of the most important zoonotic diseases, caused by invasive bacteria included in a pathogenic complex known as Leptospira Interrogans Sensu Lato [1]. This illness has a global prevalence of half million severe human cases per year [2]. The prevalence of cases worldwide is broadly underestimated, due to the variety of clinical symptoms and signs, potentially leading to misdiagnosis and undiagnosis of the disease. This zoonosis is endemic in many tropical and subtropical countries, and in the past 20 years it has emerged as a major public health problem [1].
In recent years research on immunogenic Leptospiral proteins has focused on the outer membrane proteins (OMPs) [3]. OMPs are highly conserved in most of the pathogenic serovars and induce strong immune response [4], comparable with that induced by whole cells [5]. These membrane proteins are involved in infection, transmission, virulence, survival and environmental adaptation by Leptospira. The identification of OMPs is essential for the understanding of the structure, function, interactions with the environment and the knowledge will be useful in the development of diagnostic and protective antigens for controlling the leptospirosis [6].
The design of novel diagnostic methods and vaccines for Leptospira, involves the implementation and development of new techniques and methods that allow the isolation, identification, purification and characterization of OMPs. The elucidation of the possible role of these biomolecules in the pathogenesis of leptospirosis will add up basic knowledge on biology of such pathogens.
The isolation of Leptospiral OMPs was usually carried out by extraction with detergents such as Triton X-114 and Triton X-100 [7]. Triton X-114 has been most commonly employed for the isolation of Leptospiral OMPs [8,9], whereas Triton X-100 has been used infrequently for this purpose. Matsunaga, et al. [10] fractionated Leptospiral cells using Triton X-100 for solubilization and then discarding the insoluble protoplasmic cylinder after centrifugation. A comparison of membrane protein extraction using sodium desoxycholate and Triton X-100 suggested that Triton X-100 was superior [11]. However, there are not robust evidences corroborating the usefulness of Triton X-100 as an effective detergent for Leptospiral outer membrane proteins isolation.
We describe in this research a simple and reliable method to obtain Leptospiral OMPs along with a comparison to the existing method using Triton X-114 showing the difference in phase partitioning properties of these detergents. This can be applied to the identification of novel antigenic proteins with potential to use as serodiagnostic markers and vaccine candidates.
Materials and Methods
Reference strains and culture conditions
Leptospira Interrogans serovar Copenhageni strain Fiocruz L1-130 and L. borgspetensenii serovar Castellonis strain Castellon 3 were grown in Ellinghausen, McCullough, Jonhson and Harris (EMJH) medium at 30°C and checked for the presence of contaminating aerobic bacteria after 7-10 days growth. Mid log phase cultures, identified by Petroff-Hausser counting chamber, were used for protein isolation (108 cell/ mL).
Extraction of OMPs using Triton X-100
Outer membrane protein was extracted using Triton X-100 as reported earlier [12]. Briefly, leptospires were centrifuged at 17 000×g for 30 min at 4°C and the pellets were washed twice with PBS and extracted using the buffer containing 10 mM Tris-HCl, 1 mM EDTA, 150 mMNaCl, 0.5% protease inhibitor cocktail (cat. #P8849 Sigma) and variable concentrations of Triton X-100 (0.5, 1 or 1.5%). The cells were incubated overnight at 4°C and then pelleted by centrifugation (17000 × g/ 1h/ 4°C). The protein was estimated using the BCA Protein Assay Kit and stored at -70°C in aliquots. The proteins were precipitated using Comassie Brillant Blue (CBB) as described by [13] and Trichloro Acetic Acid (TCA) as described by Mechin, et al. [14].
Extraction of outer membrane proteins with phase separation
In order to elucidate the protein extraction ability of Triton X-100 and Triton X-114, outer membrane proteins were isolated by a method described previously [15,16]. In brief, the mid log phase culture pellets were washed in PBS containing 5mM MgCl2 and then extracted in presence of either 1% Triton X-114 or 1% Triton X-100 containing 150 mM NaCl, 10 mM Tris pH 8, 1mM EDTA at 4°C. The insoluble material was removed by centrifugation at 17,000 × g for 30 min. After centrifugation 20 mM CaCl2 was added to the supernatant. Phase separation was performed by warming the supernatant to 37°C and subjecting it to centrifugation for 5min at 6,000×g. Triton X-100 phase separation was carried out at room temperature by addition of 25% NaCl. From both preparation, the detergent and aqueous phases were then separated and precipitated with acetone. The protein concentrations in aqueous and detergent phases were estimated using the BCA protein assay kit (Pierce, USA).
Protein extracts from both Triton X-100 and TritonX-114 methods were electrophoresed in 12% SDS-PAGE using the Mini-Protean® Cell II (Bio-Rad, USA) system. Used concentration of acrylamide/ bisacrylamide gel justifies the wide range of protein size to resolve (10-250KDa), as expected for Leptospiral membrane protein. After staining with silver nitrate [17], gels were scanned using a CanoScanLide 30 (USA). The comparative protein molecular mass with respect to the molecular standard was determined using UVI save D-55/20M (UVItec, England) software.
Screening for cell lysis during Triton X-100 extraction
PCR of LipL32: The absence of cell lysis after protein extraction was verified by identification of DNA using PCR. The DNA from the supernatant (centrifugation after the incubation overnight at 4°C) of Triton X-100 extraction was used to isolate DNA using High Pure PCR Template Preparation Kit for genomic DNA (ROCHE, Switzerland) and used as template to carry out PCR. The PCR amplifying a gene fragment encoding LipL32 (146bp), the major pathogenic Leptospira lipoprotein, was used as reported by Noda, et al. [18] as a marker for presence of DNA in the sample.
Zymography: A 46kDa gelatinase activity on the outer membrane [19] was used as a marker for outer membrane protein extraction. Zymography was carried out to verify the extraction of membrane protein by the presence of gelatinase activity. The sample in non-reducing SDS-PAGE sample buffer (40 mM Tris-HCl pH 6.8, 1% SDS, 2% glycerol and 0.01% bromophenol blue) was loaded without boiling to a 10% polyacrylamide gel containing 0.1% SDS and 1% gelatin. After electrophoresis, the gel was washed in 2.5% Triton X-100 solution for 30 min at room temperature followed by incubation in a reaction buffer containing 50 mM Tris-HCl pH 7.5, 1% Triton X-100 and 25mM CaCl2 at 37°C for 18h. The gels were stained using CBB solution; gelatinase activity appeared as a clear band against a blue background. The molecular masses were determined in comparison with pre-stained markers previously punched to fit the band positions using UVIsave D-55/ 20M (UVItec, England) software.
Detection of LPS in membrane protein extract
To exclude presence of LPS contamination in final protein extract, LPS detection was performed according to Riazi [20], with minor modifications. In brief, a mix of sample and proteinase K to a final concentration of 0.1 mg/ mL was prepared. The samples were incubated to 50°C for 1h and analyzed by SDS-PAGE as described above. Gels were stained with CBB and silver nitrate.
Results
Phase partitioning of protein by Triton X-100 and Triton X-114 method
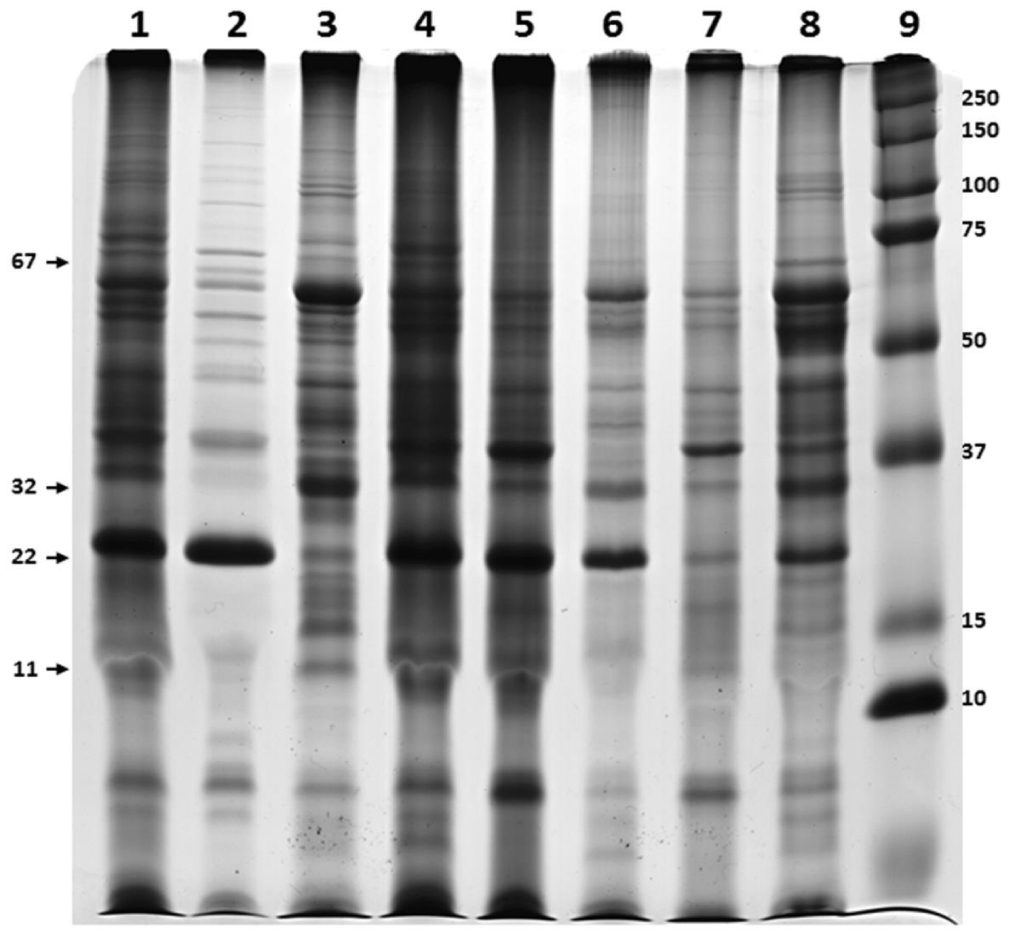
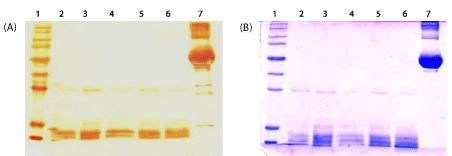
The phase partitioning properties of proteins showed that different proteins selectively enter into detergent phase of Triton X-114 or Triton X-100. For example while 67kDa and 22kDa proteins were partitioned in the detergent phase of Triton X-114, they were not retained in the detergent phase of Triton X-100. In the case of 11kDa, it was more in aqueous portion of Triton X-114 comparatively with the detergent phase of Triton X-100. In addition, 59kDa was completely partitioned into the aqueous phase of Triton X-114, it was found in both aqueous and detergent portion in the case of Triton X-100 phase partitioning as well as Triton X-114 phase partitioning of protein extracted with Triton X-100. It is also showed that Triton X-100 is more selective in retaining protein in the detergent phase as evidenced by shift of proteins from detergent to aqueous phase. None of the proteins present in Triton X-100 detergent phase changed its preference to aqueous during phase partitioning with Triton X-114 (Figure 1).
Extraction of OMPs using Triton X-100
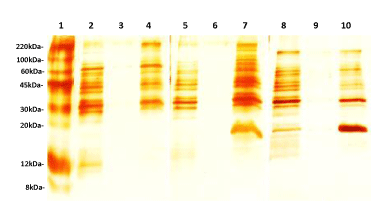
Figure 2 shows the SDS-PAGE pattern of Leptospira proteins visualized by silver staining following TCA or CBB precipitation agents at different concentrations of Triton X-100 (0.5%, 1% and 1.5%). TCA precipitation yielded a clearer and more prominent protein pattern than that obtained by the CBB method. However, omission of the precipitation step showed the best results, allowing the higher number of isolated bands in PAGE.
Twenty different bands were observed in the gel when 0.5% and 1% Triton X-100 were used, whereas 1.5% detergent yielded 21 bands (Table 1). The protein concentration obtained was 158.84 μg/ mL, 197.59 μg/ mL and 121.02 μg / mL when 0.5%, 1% and 1.5% Triton X-100 were used, respectively. A concentration of 120.34 µg/ mL was achieved for Triton X-114 extract.
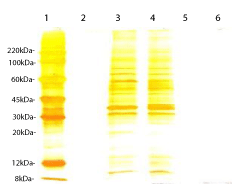
Based on the number and intensity of bands visualized by SDS-PAGE, 1.5% Triton X-100 and non-precipitation procedures were selected as optimal conditions for further experiments. The same pattern was observed when Triton X-100 and X-114 procedures were compared (Figure 3).
Screening of cell lysis during Triton X-100 extraction
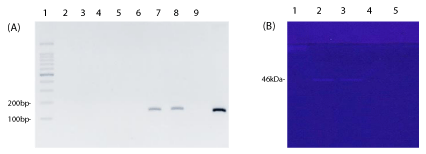
No PCR products were amplified from any of the protein extracts, in contrast to the control extract with Leptospira DNA from the extraction solution, which showed the expected amplification band (Figure 4). These results suggest the absence of cell lysis during the extraction procedure. A proteinase band of 46kDa, the Leptospira gelatinase, was visualized by the zymography analysis as an indication of extraction of outer membrane protein [18] (Figure 4).
Assessment of LPS presence in protein extract
No LPS was detected by SDS-PAGE (Figure 5). When the sample was treated with proteinase K, all bands disappeared and the CBB staining, which is more specific for protein detection than silver, not showed other electrophoretic band than that obtained with the extraction buffer and proteinase K only, suggesting LPS absence.
Discussion
Several OMPs have been identified in Leptospira, such as LigB (200kDa), LigA (130kDa), HbpA (80kDa), GspD (66,5kDa), Omp85 (60kDa), OmpL54 (54kDa), OmpL47 (47kDa), LipL46 (46kDa) or/and gelatinase (46kDa), LipL41 (41kDa), OmpL37 (37kDa), OmpL1 (33kDa), LipL32 (32kDa), Loa 22 (22kDa) and LipL21 (21kDa) [21]. Proteins located on the cell surface are of utmost importance as they are likely to be involved in the interaction of leptospires with the external environment, including host cells or extracellular matrix components. In addition, Leptospiral surface molecules may serve as targets for immune-mediated clearance [22].
This study describes the comparison of Triton X-100 and Triton X-114 as method for OMPs isolation from Leptospira with phase separation and the implementation of a reliable method particularly useful using Triton X-100. Due to the structural characteristics of Leptospira outer membrane, the use of Triton X-100 may facilitate the solublization of membrane proteins by destabilizing their interaction with the peptidoglycan layer. However, the OMPs of other bacteria, such as Escherichia coli, are tightly associated with the peptidoglycan layer, thus preventing extraction of OMPs by Triton X-100 [10].
The protein shift from detergent to aqueous phase shows that Triton X-100 is more selective in partitioning membrane proteins. The phase partitioning happened as a result of keeping the protein in the micelle form giving the hydrophobic environment similar to those found in the plasma membrane. In this case the proteins which are having more hydrophobic nature only will be partitioned with detergent phase of Triton X-100. This property may be useful to extract and selectively separate more embedded proteins of the membrane. The treatment of spirochetes with high concentration of ionic or nonionic detergents is effective in solubilizing OMPs. Nevertheless, the OMPs extraction with non-ionic detergents is better than ionic detergents, such as SDS, for as they have the potential to retain protein conformation. In addition, nonionic detergents at low concentrations, may selectively solubilize the spirochetal outer membrane, and leave the intact cytoplasmic membrane (stabilized by the cell wall). It is thought that at low concentrations the nonionic detergents Triton X-100 and Triton X-114 are able to solubilize membrane proteins. The most likely explanation is that the hydrocarbon tails of membrane proteins can be substitute for phospholipids in their interaction with membrane proteins. Definitively, the presence of cell wall is an important factor in the selectivity of nonionic detergent, which is consistent with their selective solubilization of the outer membrane of typical gram-negative bacteria [6].
As expected, SDS-PAGE gel analysis revealed that there was high correspondence among the estimated molecular sizes of the protein bands in this study with those reported for pathogenic leptospires and 1.5% of Triton X-100 was selected for further experiments since a higher number of proteins band were visualized. Future studies, will aim to resolve the identification of these proteins by mass spectrometry.
PCR analysis revealed the absence of cell lysis, suggesting that only membrane proteins were present in the extract and that the extract was free of cytoplasmic contaminants. Moreover, in the control experiment, the amplification observed with Leptospiral DNA added to extraction solution and carried out PCR confirmed the lack of PCR inhibitors in the extraction buffer. PCR can be a reliable method for cell lysis determination if appropriate controls are used to confirm the absence of PCR inhibitors from the culture medium. PCR would be useful when monoclonal antibodies for monitoring the presence of internal Leptospiral proteins are not available. Therefore, we reported here the use of PCR as alternative method for a rapid screening of Leptospiral DNA, the presence of which would indicate a condition of cell lysis.
In gelatin zymography the presence of one enzyme with gelatinase activity (46kDa), has been also reported in pathogenic leptospires [19]. In addition, other proteinases with gelatinase activity ranging in size between about 240 to 32kDa, described in L. Interrogans [23] are not found in our extraction and their cellular localization in Leptospira is also not known. Hence, our findings suggest the absence of cell lysis in protein extract with both Triton.
In conclusion, our results recommend that Triton X-100 can be used to isolate membrane proteins more specifically than Triton X-114 with no cell lysis and this method can be used in proteomic studies and study of transmembrane proteins [24].
Conflict of Interest
All authors declare to have no conflict of interest.
Acknowledgements
To Dr. Madanan G Madatriparambil, Regional Medical Research Centre (ICMR), Port Blair, India, for the critical suggestion and revision of the manuscript.
- Adler B, De la Pena Montezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010; 140: 287-296. https://goo.gl/y4B3pX
- McBride JA, Athanazio DA, Mitermayer GR, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005; 18: 376-386. https://goo.gl/pTwMwV
- Koizumi N, Watanabe H. Leptospirosis Vaccines: Past, Presentand Future. J Postgrad Med. 2005; 51: 210-214. https://goo.gl/tFBggx
- Carvalho E, Barbosa AS, Gomez RM, Cianciarullo AM, Hauk P, Abreu PA, et al. Leptospiral TlyC is an extracelular matrix-bindingproteinand does not presente hemolysin activity. FEBS Lett. 2009; 583: 1381-1385. https://goo.gl/RXMQ7Q
- Goris MG, Wagenaar JF, Hartskeerl RA, van Gorp EC, Schuller S, Monahan AM, et al. Potentinnate immune response to pathogenic Leptospira in human whole blood. PloS One. 2011; 6: 1827-1834. https://goo.gl/8CsJGw
- Cullen PA, Haake DA, Adler B. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol Ver. 2004; 28: 291-318. https://goo.gl/NbFWSw
- Nally JE, White legge JP, Carroll JA. Proteomic strategies to elucidate pathogenic mechanisms of spirochetes. Proteomics Clin Appl. 2007; 1: 1185-1197. https://goo.gl/kPu1NW
- Matsunaga J, Werneid K, Zuerner RL, Frank A, Haake DA. LipL46 is a novel surface-exposed lipoprotein expressed during Leptospiral dissemination in the mammalian host. Microbiology. 2006; 152: 3777-3786. https://goo.gl/SRSF1t
- Pinne M, Haake DA. A Comprehensive Approach to Identification of Surface-Exposed, Outer Membrane-Spanning Proteins of Leptospira Interrogans. PLoS ONE. 2009; 4: 6071-6080. https://goo.gl/DUYTGR
- Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin super family. Mol Microb. 2003; 49: 929-945. https://goo.gl/XXHbcY
- Rivera Y, Navarro Y, Mazorra Y, GonzalezA, Padron M, Valdes Y. Solubilization of components of outer membrane of Leptospira Interrogans serovars Mozdok by sodium desoxycholate and triton X-100 and evaluation of their immunogenic capacity and homologous and heterologous protection induced in hamsters. Rev Cubana Med Trop. 2005; 57: 53-54. https://goo.gl/UoUSYq
- Reyes G, Borrero R, Moya A, Padron M, Acosta M, Cabrera R, et al. Detection of adhesion proteins fibronectin and collagen present in Leptospira Interrogans serovar canicola. Vacci Monitor. 2007; 16: 1-6. https://goo.gl/CfTwd5
- Marshall T, Williams K. Two-dimensional electrophoresis of human urinary proteins following concentration by dye precipitation. Electrophoresis. 1996; 17: 1265-1272. https://goo.gl/WjZaoo
- Mechin V, Damerval C, Zivy M. Total protein extraction with TCA-acetone. Methods Mol Biol. 2007; 355: 1-8. https://goo.gl/XN7gTK
- Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B. Global Analysis of Outer Membrane Proteins from Leptospira Interrogans serovar Lai. Infect Immun.2002; 70: 2311-2318. https://goo.gl/qbrzxS
- Fricke B. Phase separation of nonionic detergents by salt addition and its application to membrane proteins. Anal Biochem.1993; 212: 154-159. https://goo.gl/KKDSCy
- Chevallet M, Luche S, Rabilloud T. Silver staining of proteins in polyacrylamide gels. Natureprotocols. 2006; 1: 1852-1858. https://goo.gl/8LPohK
- Noda AA, Rodriguez I, Rodriguez Y, Govin A, Fernandez C, Obregon AM. High sensitive PCR method for detection of pathogenic Leptospira Spp. in paraffin-embedded tissues. Rev Inst Med Trop Sao Paulo. 2014; 56: 411-415. https://goo.gl/nE6gup
- Madathiparambil MG, Cattavarayane S, Manickam GD, Singh K, Perumana SR, Sehgal SC. A Zymography Analysis of Proteinase Activity Present in Leptospira. Curr Microbiol. 2011 b; 62: 917-922. https://goo.gl/rHL7FX
- Riazi M, Abdul B, Fairuz A, Zainul FZ. A low molecular weight lipopolysaccharide antigen preparation reactive to acute leptospirosis heterologous sera. Trop Biomed. 2010; 27: 241-253. https://goo.gl/9bbDZM
- Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE. Global Proteome Analysis of Leptospira Interrogans. J Proteome Res. 2009; 8: 4564-4578. https://goo.gl/JVFiH6
- Yan Y, Chen Y, Liou W, Ding J, Chen J, Zhang J. An evaluation of the serological and epidemiological effects of the outer envelope vaccine to Leptospira. J Chin Med Assoc. 2003; 66: 224-230. https://goo.gl/eje9di
- Madathiparambil MG, Cattavarayane S, Perumana SR, Manickam GD, Sehgal SC. Presence of 46 kDa gelatinase on the outer membrane of Leptospira. Curr Microbiol. 2011 a; 62: 1478-82. https://goo.gl/4Nj1Bp
- Zor T, Selinger Z. Linearization of the Bradford Protein Assay Increases Its Sensitivity: Theoretical and Experimental Studies. Anal Biochem.1996; 236: 302–308.

Sign up for Article Alerts