Combining RPL27 with OAZ1 or RPL8 as Universal Normalizers of Gene Expression in Canine Endometrial Studies
Georg Mair1, Hermann Unger1 and Cordula Gabriel2*
1Tropical Veterinary Science Laboratory, University of Veterinary Medicine, Vienna, Austria
2Institute for Anatomy, Histology and Embryology at the Department of Pathobiology, University of Veterinary Medicine, Veterinaerplatz 1, 1210 Vienna, Austria
*Address for Correspondence: Cordula Gabriel, Institute for Anatomy, Histology and Embryology at the Department of Pathobiology, University of Veterinary Medicine, Veterinaerplatz 1, 1210 Vienna, Austria. Tel: +0043 1 250 77 3403; E-Mail: cordula.gabriel@vetmeduni.ac.at
Submitted: 19 July 2017; Approved: 20 September 2017; Published: 22 September 2017
Citation this article: Mair G, Unger H, Gabriel C. Combining RPL27 with OAZ1 or RPL8 as Universal Normalizers of Gene Expression in Canine Endometrial Studies. Int J Vet Sci Technol. 2017;1(1): 023-034.
Copyright: © 2017 Gabriel C, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Keywords: Laser Capture Micro dissection (LCM); Reference genes; Endometrium; Pyometra
Download Fulltext PDF
Background: The canine endometrium represents a complex, heterogeneous tissue with several cell populations featuring divergent gene expression patterns that become of high interest during cyclic hormonal stimulation, or in response to pathogen-activated immunological activation like in the case of pyometra. Assuming that reference gene expression stability is not shared between the different endometrial cell types, a whole tissue-based approach may not be adequate. Dependable qRT-PCR quantification of endo- or exogenously induced transcript me changes is primarily reinforced by normalisation with reference genes. Thorough evaluation of these endogenous controls is important, but has so far been insufficiently addressed, especially in reproductive veterinary medicine. Often, reference gene selection relies on results obtained with other species, or considers only tissue and not cell-specific results.
Results: We evaluated the stability of nine putative reference genes in healthy and pyometra-affected canine uterine tissue homogenates, and the respective endometrial cell types. For cell-specific analysis endometrial stromal and epithelial cells were either isolated via Laser-Capture Micro dissection (LCM) from tissue cryo sections or cultivated under hormonal stimulation in vitro. While single-gene stability ranking did not present a universal solution, the inclusion of arbitrary normalizers composed of the geometric mean of paired genes computed by group-specific Norm Finder analysis showed that using RPL27 combined with either RPL8 or OAZ1 resulted in remarkable stability values. Both arbitrary reference genes were top-ranked in the tissue and cell culture data sets, and were useful especially for the highly variable LCM samples.
Conclusion: The combination of RPL27 with either RPL8 or OAZ1 was successfully identified as best working normalizer in gene expression data acquisition of different sample types of the canine endometrium.
Abbreviations
ACTB: β-Actin; Cq: Quantitation cycle; CT: Cycle treshold; E: Estrogen; EECs: Endometrial Epithelial Cells; ESCs: Endometrial Stromal Cells; GAPDH: Glyceraldehyde-3-Phosphate Dehydrogenase; GUSB: β-Glucuronidase; HPRT: Hypoxanthine-Guanine Phospho Ribosyl Transferase; LCM: Laser Capture Microdissection; OAZ1: Ornithine decarboxylase antizyme 1; P: Progesterone; RPL27: Ribosomal protein L27; RPL32: Ribosomal protein L32; RPL8: Ribosomal protein L8; RPS20: Ribosomal protein S20
Introduction
Molecular research determining physiological as well as pathological changes in the uterus is faced with the lack of universal reference genes to determine relative changes of gene induction and expression. The complexity of interacting cell types in the uterine wall together with hormone-driven cyclic changes affects interpretation of expression data regardless of whether total uterine homogenates, layer-specific samples or isolated cell populations are being analyzed [1]. Reference genes selected for normalizing such expression data are likewise restricted to a cell type-specific application, in contrast to normalizers defined by whole-tissue analysis. This is evident from different studies revealing divergent transcript me profiles closely related to the day of the sexual cycle after separating endometrial and stromal fractions by laser capture micro dissection (LCM) [2-4]. Although specific guidelines for the basics in quantitative RT-PCR analyses have been available since 2009 [5-7], only a few studies in veterinary medicine approached the evaluation of suitable reference genes [8-10]. In particular, pathological effects in different layers of the uterus, and immune system driven influences are essentially unexplored.
Pyometra, the purulent inflammation of the uterus, is a frequently occurring pathology of the canine endometrium in adult intact bitches, and is thus of major interest in veterinary medicine and reproduction [11,12]. In six out of ten cases the infection progresses into a potentially life-threatening systemic inflammation [13]. Characterization of relevant genetic markers involved in the development of this pathology thus has a high clinical relevance. A detailed analysis of the genetic profiles of affected and healthy uteri already identified more than 800 genes as being up regulated in pyometra-affected uteri [14]. These results reflect the serious alterations observed in the canine uterine tissue during pyometra and the associated inflammatory response of the endometrium. Transcriptome studies concerning canine pyometra development and etiology are however rare, although a detailed investigation of relevant marker genes is considered necessary [15-18]. Moreover, abundant qRT-PCR data were produced without assessing suitable reference genes essential for the normalization of these data. So far, TUBA1A (α-tubulin) as an internal standard [15], as well as RPL27 [17,18] and OAZ1 [16] have been applied in the context of the canine pyometra-affected endometrium. OAZ1 and RPL27 were equally identified as stable reference genes in the cyclic porcine uterus [19], but their efficiency for specifically normalizing profiles of the different cell populations in the canine uterus has never been demonstrated.
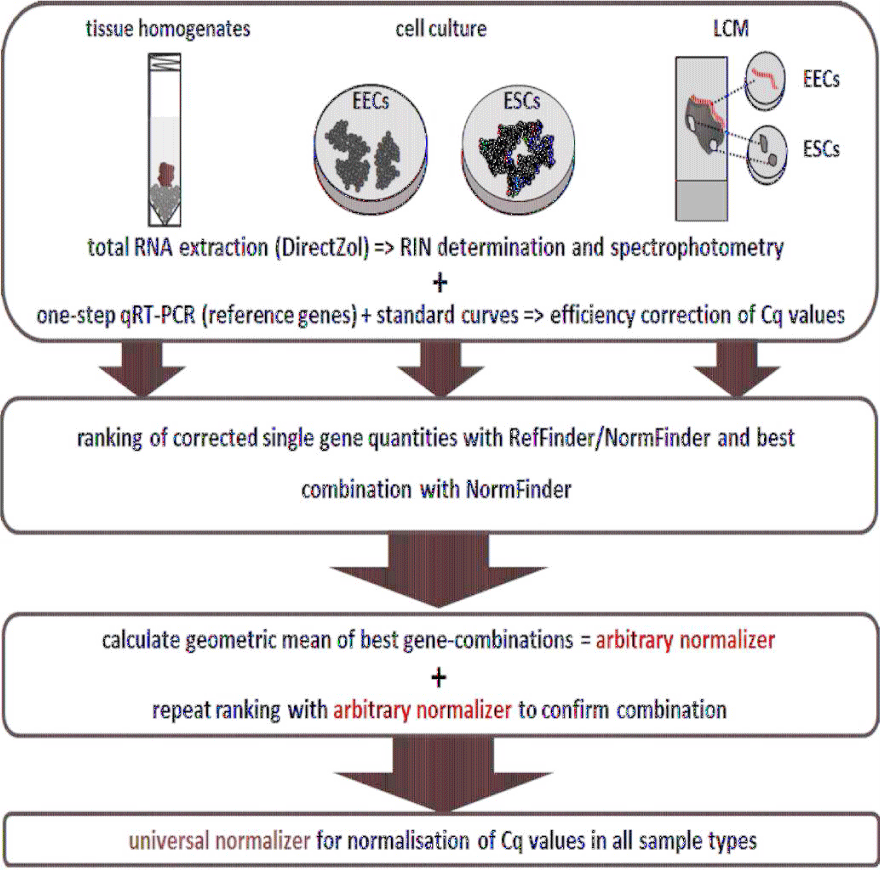
We thus developed a stringent workflow for evaluating reference genes applicable for healthy and pyometra-affected canine uteri homogenates and isolated uterine epithelial and stromal cell populations. Here, the influence of cyclic changes was addressed by treating these cultured cells with estrogen and progesterone. We analyzed the nine most promising genes not affected by pyometra [14], under the assumption that potentially the mean of two or more genes could present an arbitrary normalizer.
We therefore employed healthy and pyometra-affected uterine tissue samples, two different endometrial cell types stimulated with steroid hormones in vitro and LCM samples of the respective endometrial cell types, for reference gene analysis and quantification of their respective expression levels. A mathematical and statistical approach was taken to evaluate the optimal combination of genes and their expression levels for their use as reference genes in quantitative, cell-specific profile studies of the canine endometrium.
Materials and Methods
Tissue and Cell Preparation
Uterine tissue was collected from routine ovariohysterectomy (n = 3) and surgical pyometra treatment (n = 3). Surgery was performed under general anesthesia at the Department of Companion Animals and Horses, Section of Obstetrics, Gynecology and Andrology of the University of Veterinary Medicine at Vienna, Austria. Tissue sampling and evaluation as well as anonymized publication of the received data were in accordance with the pet owners and the project was approved by the local ethical commission at the Vetmeduni Vienna, Austria to be based on the respective regulations of good scientific practice. The dissected uterine tracts were transported in sterile Dulbecco’s Phosphate Buffered Saline (Sigma-Aldrich, Steinheim, Germany) containing 0.5% Gentamicin (Biochrom, Berlin, Germany) and 1.5% Nystatin suspension (10,000 units/mL in DPBS; Sigma-Aldrich), at 4-8°C.
For the histological evaluation, samples (1 cm³) of the uterine horns (cranial and intermediate regions) as well as of the uterine body (bifurcation) were separated and immersion-fixed in 4% buffered formaldehyde for 24 to 48 hr at 4°C, and then embedded in Paraplast® (Vogel, Giessen, Germany). For histological evaluation, sections of 3 µm thickness were cut and stained with Hematoxylin and Eosin (H&E) according to Romeis [20]. Evaluation of the sections was performed by light microscopy using a Polyvar microscope (Reichert-Jung, Vienna, Austria) connected to a DS-Fi1 digital camera (Nikon, Vienna, Austria) with Nikon NIS-Elements software.
For the mRNA analyses of tissue homogenates of the same regions of three healthy and the three pyometra-affected uteri, these samples were snap-frozen in 500µL RNA later (Qiagen, Hilden, Germany) in liquid nitrogen and stored at -80°C. For the mRNA analyses of the LCM samples, uterine samples (n = 3, healthy) of the mentioned regions were embedded in Tissue-Tek® O.C.T™ (Sanova Pharma GmbH, Vienna, Austria), and cryotome sections of 10µm thickness were cut with Leica Cryocut CM 1800 at -16°C.
Laser capture microdissection (LCM)
For the LCM samples, cryotome sections were placed on a MMI Membrane Slide with a 1.4 µm PET membrane (Molecular Machines & Industries, Glattbrugg, Switzerland). Cryotome sections were air dried and fixed for 30 s in 70% ethanol, subsequently rehydrated for 30 s in aqua dest and stained for 20 s with HistoGeneTM LCM Frozen Section Staining Kit (Arcturus, CA, USA) followed by washing in aqua dest for 30 s. After dehydration (30 s per step: 70%, 80%, 90%, 100% ethanol, 5 min xylol) membrane slides were air-dried for 5 min and placed on RNAse-free glass slides. Per sample, two slides with four to six tissue sections and six CapSure® Macro LCM caps (Arcturus, CA, USA) were placed into the VERITAS ® Laser Micro dissector. Endometrial Epithelial Cells (EECs) and Stromal Cells (ESCs) were separately localized (20x magnification) and marked for laser capturing. The respective selected cell population was cut and captured with two different lasers. The laser power setting ranged from 60 to 80 mw, pulse settings from 1700 to 2500 μs. The spot size for fixing the cells onto the caps was determined prior to micro dissection, and adapted to each cell type [21].
Cell culture
For mRNA analysis of the uterine cell populations in vitro the endometrium was separated from the myometrium of a healthy anestrous uterus. The two different Endometrial Cell Types (ESCs and EECs) were isolated as previously published [22]. In brief, after digestion of the endometrium in standard medium (88% M199 with L-glutamine (HyClone Laboratories, South Logan, Utah, USA); 1% antibiotic-antimycotic solution (PAA Laboratories, Pasching, Austria); 1% Fungizone®, liquid (Gibco, Thermo Fisher Scientific, Austria), without fetal calf serum, FCS) containing 1% collagenase type I (Sigma-Aldrich) for 3 hr, the epithelial structures (surface epithelium and glandular epithelium) were separated via two consecutive filtrations steps (280 µm and 40 µm mesh size, respectively; BD Biosciences, Erembodegem, Belgium). The suspension of ESCs was immediately centrifuged (2000 rpm, 2 min) and resuspended in fresh standard medium containing 10% FCS (Sigma-Aldrich). The ESCs were cultured in 25 cm² cell culture flasks (Sarstedt, Newton, USA) with standard medium containing 10% FCS for 5 days at 37°C and 5% CO2. Epithelial structures were removed from the filter membrane, and transferred into a petri dish containing standard medium, and then collected in a 15 mL tube for centrifugation (2 min, 2000 rpm). The structures were resuspended with 1% trypsin/EDTA (Biochrom, Berlin, Germany) for 4 min after two wash steps with PBS, and subsequently centrifuged for 2 min (2000 rpm) to remove trypsin/EDTA. Dissection of the remaining structures in the pellet followed on resuspension in 3 mL standard medium (10% FCS) and was supported mechanically via pipetting for a single cell suspension of EECs. The EECs were cultured in 25-cm² flasks at 37°C and 5% CO2 for seven days with additional 1% trypsin/EDTA treatment on days five and seven for transfer to a 75-cm² cell culture flask (Sarstedt, Newton, USA). For each cell suspension 1 mL was used to produce four glass platelets (24mm, VWR, Vienna, Austria) (each with 250 µL cell suspension and 750 µL standard medium with 10% FCS) in a 24-well plate for immunohistochemical analysis of the isolated cell type. Cells were grown on the 75-cm² flasks for four (ESCs) to eight (EECs) days to reach a confluence of 80%. Then cells were stimulated with either 15 pg/mL 17ß-estradiol (E) or 15 ng / mL progesterone (P) for 48 hr or cultured as control group without stimulation. The cells were scraped with a cell scraper (Greiner Bio-One, Kremsmunster, Austria) within the medium, and transferred to a 15-mL tube. After centrifugation and decantation the dry cell pellets were frozen at -80°C. The cell culture experiment was repeated in two independent approaches for each cell type.
Immunohistochemistry
Identification of the respective cell type was performed by means of immunohistochemistry. Cells grown to 80% confluence on the glass platelets were washed two times with PBS and then fixed with 4% buffered formalin for 10 min at room temperature. Afterwards, the formalin was removed and the glass platelets were washed two times with aqua dest followed by overnight drying at room temperature. Glass platelets were subsequently fixed on slides with “DPX new” (Merck Millipore, Merck Chemicals and Life Science, Vienna, Austria) for staining on the following day. EECs were identified with an anti-cytokeratin antibody (mouse clone AE1 + AE3; Sigma-Aldrich, dilution 1:250) whereas ESCs were specified with an anti-vimentin antibody (mouse clone V9, Dako, Agilent Technologies, dilution 1:200). Cells on the slides were permeabilized with 0.15% Triton X-100 in PBS for 15 min at 4°C and then washed three times in PBS. After a peroxidase block, the slides were incubated in 1.5% normal goat serum (Vector Laboratories, Burlingame, CA) for 30 min to minimize unspecific binding of the primary antibody. For anti-vimentin antibody boiling in citrate buffer (pH 6.0, 2 x 5 min), and for the anti-cytokeratin antibody boiling in Tris-EDTA (pH 9.0, 3 x 5 min) was performed. Primary antibodies were incubated over night at 4°C and detected by a fluorescent secondary antibody (Alexa Fluor 488, Molecular Probes, Thermo Fisher Scientific, dilution 1:100) and 4’,6-Diamidino-2-Phenylindole (DAPI, Molecular Probes, Thermo Fisher Scientific) was used for nuclear counterstaining. Evaluation of the sections was performed using confocal laser scanning microscopy (Zeiss, LSM 510 Meta, Vienna, Austria) with ZEN2000 software (Zeiss, Vienna, Austria).
RNA treatment
Whole-tissue biopsies (3 mm³) were homogenized in 400 µl QIAzol reagent (Qiagen) and with a maximum of 70 standard ceramic beads using the MagNA Lyser instrument (Roche Life Science, Vienna, Austria). A portion of the homogenate (350 µL) was mixed with an equal volume of ethanol (99%), and total RNA was extracted following the Zymo Direct-zol kit (Zymo Research Corp., Irvine, USA) protocol. Cultured stromal and epithelial cells were pelleted and re-suspended with 350 µl of QIAzol reagent before lysis for 15 min at 37°C in a heat shaker and incubation at room temperature for another 15 min before adding an equal volume of ethanol. Lysis of the LCM samples was achieved by adding 100 µl QIAzol to the dissected cells glued on the recovery caps, and incubation for 45 min at 37°C, then 15 min at room temperature. A further 100 µl of QIAzol were added to the homogenate before mixing with an equal volume of ethanol.
RNA quality was analyzed for the whole tissue and the cell culture samples using the RNA 6000 Nano kit and the 2100 Bio analyzer instrument (Agilent Technologies, Santa Clara, USA) determining a mean RNA integrity number (RIN) of 8.1. The total RNA amount was further determined using the Nano Drop 2000c platform (Thermo Fisher Scientific).
Primer design
Peer-reviewed primer sets for nine candidate genes (Table 1) were tested for specificity, temperature and dimerization using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) [23] and Net Primer (http://www.premierbiosoft.com/netprimer/). New sets were designed using either Primer-BLAST or the Perl Primer tool (http://perlprimer.sourceforge.net/) [24], limiting the output by temperature, amplicon length and large intron spanning exon-exon placement. Final set definition was achieved by testing for dimerization and hairpin likelihood with the Net Primer tool and for amplicon secondary structures using the Mfold algorithm (http://unafold.rna.albany.edu/?q=mfold/download-mfold) [25] for DNA with setup-dependent settings for temperature and salt correction.
qRT-PCR
The gene-specific one-step qRT-PCR approaches were performed in a 10 µl final reaction volume and consisted of 5 µl RevTrans Mastermix (2x) with the ds DNA binding dye Eva Green (Bio & SELL, Feucht, Germany), 200 nM of each primer plus a volume of water sufficient to fill up to the 8 µl master mix volume per reaction. Total RNA was added with a final volume of 2 µL, whereas the final RNA concentration of measured samples was 10 ng.
Amplification was performed using the Viia 7 384-well (Thermo Fisher Scientific) and the Rotor-Gene Q 72-well platform (Qiagen) and included a preceding reverse transcription step at 50°C for 15 min, followed by heat denaturation at 95°C for 5 min, 45 cycles with denaturation at 95°C for 15 s and annealing/elongation at 60°C for 50 s and a melting curve step. The efficiency of amplification was determined by standard curve assay using pooled total RNA and the LinReg PCR software [27], from which Cycle of quantification (Cq) values for each gene were derived.
Reference gene stability determination
Stability of expression across tissue- and cell-type specific data sets was assessed using the efficiency-corrected quantities of the reference genes in the calculation matrix of the online tool Ref Finder (http://omictools.com/reffinder-tool)and the Norm Finder Excel add-in [28]. Ref Finder computed a comprehensive ranking of the genes after analysis with the statistical algorithms Best Keeper [29], ge Norm [30] and Norm Finder [28] and the ΔCq method. The stand-alone Norm Finder tool additionally defined a best combination of two genes after definition of group identifiers. The averaged quantities of these combinations were included as arbitrary reference genes (arbitrary normalizers, respectively) in a final stability assessment with the Ref Finder tool.
Results
Workflow to establish optimal reference genes for expression studies
In order to establish a set of reference genes for cell-specific gene profiling studies of the canine endometrium, we established a workflow that involved tissue and cell selection, gene expression measurement, and data analysis (Figure 1). Firstly, when tissue samples were the source material, these were subjected to detailed histological examination to assess whether they were healthy or showed signs indicative of pathologies. Then, total RNA was extracted from tissue homogenates or cultured cells, and used as a template for one-step qRT-PCRs to quantitatively evaluate the expression of nine genes: ACTB (β-actin), GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase), GUSB (β-Glucuronidase), HPRT1 (Hypoxanthine-Guanine Phosphoribosyltransferase), OAZ1 (Ornithine Decarboxylase Antizyme 1), RPL8 (Ribosomal Protein L8), RPL27 (Ribosomal Protein L27), RPL32 (Ribosomal Protein L32), and RPS20 (Ribosomal Protein S20). Data from the qRT-PCR measurements were then subjected to analysis with various algorithms in order to rank and assess the suitability of each gene as a reference.
Healthy and pyometra-affected uteri
The first source of biological material that we used to define optimum reference genes was complete canine uterus, either healthy or pyometra-affected. Histological evaluation of the healthy (control) group of uteri showed these to be without any pathological indications (Supplementary File 1A). In the pyometra-affected group the surface and the crypt epithelium displayed a foamy appearance and were detached from the stroma in some regions (Supplementary File1B). The stroma had partially a loose appearance, which might result from the presence of edema fluid. In the dense stromal regions there was a large number of neutrophils and macrophages, and aggregates of plasma cells.
Tissue homogenates of healthy (n = 3) and pyometra-affected (n = 3) uteri were used as source material for RNA extraction and qRT-PCR-based measurement of the expression of the nine candidate genes. Expression data obtained from these aggregated cell-types were used to rank the putative reference genes. The correlation between the gene data sets was assessed to determine the variation due to either random technical errors (e.g. cDNA synthesis) or influences that affected all genes equivalently (e.g. sample quality). The expression data were confirmed as being reliable for further reference gene specific calculations, demonstrating that no correlation was given between the different assays (data not shown).
Stability in these reference gene sets was first determined with Ref Finder, which combines the out puts from the most commonly-applied stability calculations. Briefly, GAPDH was defined as best single normalizer by the mean of all algorithms, whereas ACTB was the least reliable option. The highly-expressed RPL genes were top-ranked for variation analysis by Best Keeper, but at middle positions in the ranking summary. The Norm Finder calculation of the Ref Finder matrix top-ranked GAPDH without log-transformation and grouping of the raw Cq values (Table 2).
Using the stand-alone Norm Finder Excel add-on, the values were subsequently log transformed and grouped to analyze inter-group variance and define the combined normalizer (Table 3). Using log-transformed but ungrouped values OAZ1 achieved the top-ranking. Defining a healthy and a pyometra-affected group for the inter-group variance parameter again changed the ranking. GAPDH was identified as the best single gene independent of the log transformation. The best combined normalizer for analysis of healthy and pyometra-affected canine uterine tissue was the combination of RPL8 and RPL27, equally with or without log transformation.
Uterine cell populations in vitro
Secondly, we sought to establish optimal reference genes in cultured cells corresponding to specific uterus-derived cell lineages. Cells isolated from healthy canine uteri (n = 3) and cultured as Endometrial Epithelial Cells (EECs) were identified via immunohistochemical-positive staining for cytokeratin, and negative staining for the mesenchymal marker vimentin (Supplementary File 2A). Cultured Endometrial Stromal Cells (ESCs) isolated and cultured from the same tissue samples were positive for vimentin and negative for the epithelial cytoskeletal filament cytokeratin (Supplementary File 2B). The population doubling time for the EECs was 72-96 hr, whereas that for the ESCs was less than 48 hr. The two endometrial cell types were subsequently stimulated with either estrogen or progesterone for 48 hr to introduce an additional factor for stability determination of the selected reference genes.
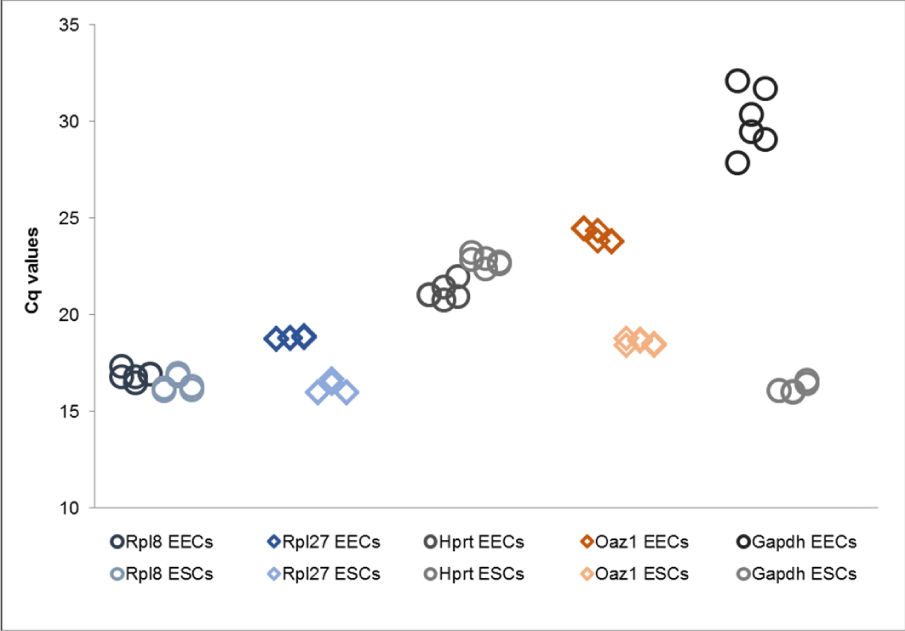
Interestingly, assembling a combined data set of both cell types for correlation and comprehensive RefFinder analysis illustrated that gene expression ratios differed considerably between EECs and ESCs. The expression levels of several genes were considerably reduced in EECs, with GAPDH being detected with a high Cq value almost at the end of the dynamic range of the assay. The expression levels of the RPL genes in ESCs, however, were similar to the levels in ESCs. As a consequence these genes were top-ranked by comprehensive Ref Finder analysis due to the lower expression difference (Table 4). Cell type-specific analysis in contrast first ranked OAZ1 for the ESCs and RPL32 for the EEC data set by the mean of all algorithms. GAPDH was considered the least appropriate reference gene for combined and separate cell-type analysis (Table 4). The Norm Finder add-on identified RPL27 as the most stable gene of the combined data sets without, and OAZ1 as the stablest gene with log transformation. Separate analysis of the cell-type data sets selected RPL20 for EECs and OAZ1 for ESCs, irrespective of the log transformation. Stratifying the combined data sets for the cell types to apply the intergroup analysis confirmed OAZ1 as being the most stable gene and best normalizer if fused with RPL27. Calculations with the combined sets being partitioned for no treatment or estrogen and progesterone stimulation selected the same fused normalizer and RPL27 as best gene. Using the equal partition for each cell type in separate analysis again identified RPS20 for the EECs and OAZ1 for the ESCs as best genes. The best normalizer for the EECs was the combination of RPS20 and RPL27, while it was OAZ1 and RPL32 for the ESCs). Analysis of variance and ΔCq definition finally demonstrated that the stimulation with estrogen or progesterone did not interfere with the stability of any gene, regardless of the cell type (data not shown).
Proof of principle analysis of suggested gene combinations as arbitrary normalizers
Two of the best combinations computed by Norm Finder for the tissue and the cell culture experiments after setting respective group identifiers were included in a final ranking analysis with the Ref Finder tool. This proof-of-principle analysis demonstrated that the suggested genes-if averaged and used as arbitrary normalizers - were assessed as having greater stability compared to the single-gene solutions. Both normalizers thus were predominately top-ranked by the applied algorithms independent of the respective data sets (Table 5,6).
If not top-ranked, at least one of the combined genes was among the two top candidates. This indicated the potential of the arbitrary normalizers to allow for an unchanged normalization strategy equivalent for all sample types in pyometra-related studies, in contrast to the use of different single genes. A comprehensive ranking of all positions for the RPL8/RPL27 and the OAZ1/RPL27 normalizer finally top-ranked the latter combination as a reliable reference gene solution (Table 7, Figure 2).
Uterine cell populations isolated by LCM
The third source of biological material we used to assess the putative reference genes were cell type-specific sections of canine uterus excised by LCM. Here, samples of the canine endometria selected for LCM varied in size between 800 000 µm² (surface and glandular EECs, Supplementary File 3A) and 300000 µm² (ESCs, Supplementary File 3B), due to existent lumina and necessary safety distance areas for cutting surface and glandular endometrial cells. The real areas of EECs were measured before selection of the cutting areas, and varied between 47000 and 56000 µm².
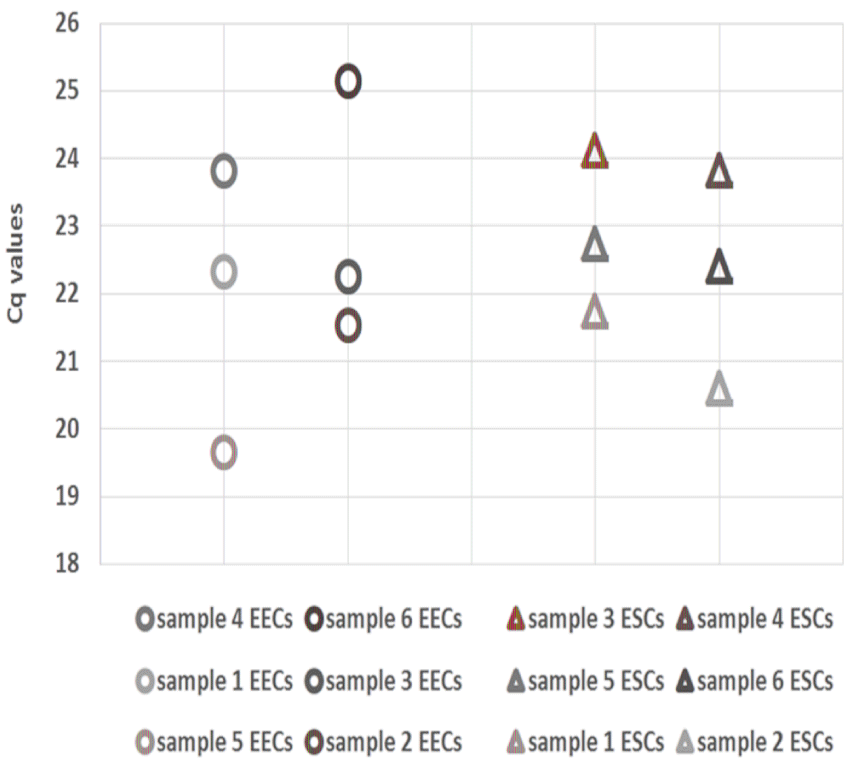
Only individual captures were considered for gene quantification in LCM samples. Pooling captures of different spots was avoided, to allow a more marked description of technical variation. For LCM experiments not only RNA measurement mistakes or varying RT efficiency need consideration, but also the laser treatment and resulting damage to the outer cells, the handling time, recovery, and undetected additional layers, all of which can cause non-biological influences. The observed inter-sample variance in EECs and ESCs samples of one of the top candidate reference genes, RPL27, reflected these concerns (Figure 3 and Supplementary File 4).
The Cq values, as judged by eyes, were less scattered in the EECs with however an inconsistent variance pattern of the samples for the different gene assays. The scattering of the Cq values was higher in the ESCs samples while the variance pattern was more consistent. Another factor was an observed deviation between the highly-expressed and medium-expressed genes. Due to the reduced amount of RNA per reaction, the scattering was increased for the latter genes. This confirmed first of all the challenge of using LCM samples for reference gene evaluation, as the data were not useful for a computed stability analysis. We however used the experiment for testing the most consistent and unbiased description of non-biological influences, and finally analyzed the artificial variance pattern in the defined normalizers. Using the single-gene results as a benchmark, the combination of RPL27/OAZ1 was specified as the best option for gene-of-interest normalization regarding the influence of the scattering by different expression levels combined with the inherent variance in a sample set. A combination of RPL27/RPL8 was considered more likely to convey the pattern exclusively of the highly-expressed genes, thus reducing the variance evident in other gene assays. The remaining genes HPRT1 and GAPDH were not considered because HPRT1 is known to be more likely influenced by biological factors [31] (and showed the most variance), and GAPDH expression was more scattered in the ESC samples compared to the other genes.
Discussion
Identification and application of reference gene sets is essential for presenting gene expression data in a meaningful way. Normalizing the raw expression values of genes of interest reduces technical influences caused by varying reverse transcription efficiencies or template concentrations. Reference genes have to be determined in a way such that they are unaffected by the respective study design and the cell types involved, to avoid affection by different cell type quantities.
These aspects present a specific challenge for analyzing reproductive tissues such as the endometrium, which is a highly dynamic tissue undergoing serious cyclic changes (for review, see [1,32]). Defining a putative normalization strategy is confronted with difficulties associated with changes in growth, differentiation, desquamation and regeneration under steroid hormone as well as cytokine/chemokine influences. The composition of the endometrium is another factor as it includes epithelial and stromal cells as major cell types, which interact closely but have to serve different tasks [33,34]. Performing a disease-related evaluation of putative reference genes with uterine tissue samples is thus technically useful and often necessary, but more a starting point considering tissue heterogeneity and cell-specific reference gene evaluation.
In a tissue homogenate the effects on a specific cell type may be concealed if this cell type comprises a small or the smallest proportion, which was the case for the endometrial epithelial cells in the uterine homogenates applied for the present study. Uterine tissue comprises only a small percentage of glandular and surface epithelial cells that nevertheless line the mucosal surface as a first line of defense [35]. As an efficient physical barrier against infection, epithelial cells have evolved innate immune antimicrobial functions and can modulate the recruitment and activity of immune cells by chemokine release [36,37]. Therefore, studies concerning pyometra etiology have to focus on the epithelial cells, but gene expression analyses of tissue homogenates will mainly generate patterns that result from the predominant stromal cells.
Human studies disclosed the importance of cell-specific gene expression profiling by separating endometrial epithelial and stromal cell fractions using LCM [2,4,38], and assessing day-specific mRNA quantitation during the estrous cycle [4,38]. Including LCM in a study design is indeed a beneficial strategy to recover cell layers or small amounts of specific cells out of a heterologous tissue sample [39], while alternative applications such as cell isolation by FACS [40] need critical consideration (for review [41]). LCM-based isolation of cell samples however has the consequences of quality loss because of the laser treatment, unpredictable influences on recovery by the adhesive surface of the capture caps, and the small scale of target cells in the case of un-pooled samples. These factors cause high variation within a sample set, and typically lead to the need for a reliable normalizer for other sample types. Therefore we likewise started with whole-tissue samples for the development of a rigorous normalization strategy of gene expression profiles in healthy and pyometra-affected canine uterine samples and isolated endometrial cells. A subsequent consideration of steroid- and untreated endometrial cells and LCM samples involved address sing the issues of cell-type specificity and the possible effects of hormonal stimulation. A subsequent consideration of cell-type specificity was performed by the inclusion of an LCM-isolated endometrial cell population. The same cell populations were isolated from the endometrium, and cultured to determine the influences of estrogen and progesterone on cell-type specific reference gene expression patterns.
The approach faced that challenge that only a small number of previously-evaluated candidate reference genes had been described for canine expression studies. Equally valuable canine-based microarray data are also rare, in contrast to those from human or murine studies. Therefore, highly expressed RPL genes, the frequently-communicated OAZ1 and “traditional” reference genes were included, with one of the “traditional” genes, GAPDH, being top-ranked in the initial analysis of the whole tissue data set. This was (strictly speaking) reasonable considering its abundantly discussed history as a housekeeping gene, being expressed at equivalent levels in different organs or cell types. It was indeed interesting to see the highly expressed RPL genes not to be top-ranked, although the Cq values per sample and hence the overall expression data was more consistent. This homogeneity may likewise be considered to equally indicate high expression stability. Instead, more variant, mid-level expressed genes were better positioned. Considering that the presence of variance is generally due to technical reasons in homogenous sample sets, the mid-level expressed genes may better convey means of sample quality or metabolic differences, factors that are certainly considered by our stability prediction methods.
Metabolic differences affecting the expression patterns were demonstrated after the in vitro experiments using cultivated endometrial cells. The expression ratios in the epithelial cell populations were significantly reduced compared to stromal cell populations, except for the RPL genes and HPRT1. Surprisingly, GAPDH was expressed only at a basal level in the cultivated epithelial cells. The less imbalanced RPL genes were hence at the top-ranked positions within the (however only partly applicable) combined data set. Cell-specific analysis confirmed this ranking just for the epithelial cells, but top-ranked OAZ1 for the stromal cells. Stimulation with estrogen and progesterone did not affect the top candidates, pointing to a valuable applicability of these genes for studying cyclic changes of the investigated endometrial cell types. GAPDH, amongst or the least stable candidate reference genes for the canine endometrial in vitro experiments, was in contrast rejected as a candidate although it was considered the best for the whole-tissue experiment. A study involving the estrogen and progesterone stimulation of primary cultivated human thyroid cells demonstrated comparable results using ACTB as the reference gene of choice in the stimulation assay when compared to GAPDH and others [42]. The ACTB gene however was not a suitable reference gene in the present study using canine tissue and cells. This demonstrates convincingly that reference genes described in studies involving other species should be treated as putative and need further evaluation and validation before use in studies involving the species of interest.
Here, the Ref Finder tool was highly versatile for ranking the genes, as it included the most respected algorithms. Plain rankings of the candidate genes nevertheless did not completely fulfill our demand for a potentially universal normalization strategy in our pyometra-related study. The tool lacked the determination of the best gene combination, as computable using the stand-alone Norm Finder algorithm. The best combination may not necessarily include the top-ranked genes, but comprises genes that theoretically result in a top stability for normalization, taking into account different aspects of a study design, after averaging the quantified values as an arbitrary normalizer. We challenged this by including two arbitrary normalizers for either data set in another ranking with OAZ1/RPL27 finally top-ranked independent of the sample type. Top-positioning of this normalizer was accepted to be caused by the highest stability and thus allows for a reliable and universal normalization strategy of future pyometra experiments. Our results furthermore are partially consistent with other studies using RPL27 as a reference gene in canine pyometra tissue homogenates [17,18], and a study concerning estrous cycle-related changes in the canine endometrium, which determined OAZ1 as the most versatile reference gene [16]. Its usability for our in vitro stimulation assays confirms OAZ1 as a suitable reference gene for analyzing cyclic changes in the canine endometrium (more specifically the stromal cells).
A single or stand-alone reference gene however is not an adequate normalization strategy [30,43]. At least two genes should be defined according to past reports, a prerequisite addressed in this study by the defined normalizers RPL8/RPL27 and OAZ1/RPL27. The latter combination was actually reported as the best normalization strategy for tissue homogenates of the ovary, oviduct and uterus of pre-pubertal and pubertal gilts of different breeds and different stages of the estrous cycle, but no computed confirmation was included for the averaged normalizer [19]. In this study, we averaged not only the determined endogenous genes but ranked them again with the other candidate genes. This successfully determined the inherent stability of the combinations independent of the working hypothesis or the sample type as described above.
The last target, testing the genes in LCM experiments, was a serious challenge for this strategy. The gene-specific expression patterns determined with the individual samples were distinctly affected by non-biological influences - although the RNA extraction workflow was additionally optimized- and in the case of the EECs inconsistent. The inconsistent patterns and the consequential influence of the expression level on scattering of the Cq values caused us to recommend the combination of the highly-expressed (RPL27) and medium-expressed (OAZ1) genes. It was evident that the use of this normalizer did not outbalance and thus hide technical variance. To be more precise, we already demonstrated the high stability of the selected normalizer for different sample types. This allowed the role of the normalizer to be defined in the case of LCM experiments for the correct depiction of the variance for high-, medium- and low-expressed genes before curating the raw quantities. This was more evident for the RPL27/OAZ1 combination than for the combination of two highly-expressed genes. Still, the discussed aspects reduced the validity of a computed reference gene stability-ranking, and illustrate why such an approach is rarely discussed at all. The intrinsic advantage of this technique – analyzing selected cell types in their natural habitat – on the other hand is crucial for an understanding in gene expression of both physiological and pathological alterations (for review [39]). Without adequate normalization tools, determining the impact of cell environment including both, cell-cell and cell-extracellular matrix interactions, on gene expression is otherwise not achievable.
We successfully defined such a reliable normalization strategy by demonstrating the arbitrary combination of OAZ1 or RPL8 and RPL27 as a versatile, unique normalizer for experiments with canine uterine tissue homogenates, steroid- or un-stimulated endometrial cultured cells and even LCM-captured endometrial cell fractions. This optimal case – a universal normalization solution for all sample types – minimizes additional optimization effort and variation by systematically changing the reference genes for different sample types. The mostly top-ranked gene OAZ1 was additionally highlighted as being optimal for the cell culture-related experiments, and the highly-expressed gene RPL27 was shown to be versatile even for different sample types. Regarding the potential application of these assays for pyometra-affected uteri, the use of these universal normalizers provides opportunities for researchers to explore a wide range of potential etiological mechanisms, from hormone-driven overreactions, to bacterial invaders. Our findings may provide support for future canine studies using the reference genes characterized here, and our strategy for defining highly stable normalizer combinations may prove useful to other groups wishing to define optimum reference genes for their expression studies.
Acknowledgements
We would like to thank Irina Kolarov for her assistance with the cell culture experiments and processing the preparatory work for the qRT-PCR analyses, Claudia Hochsmann for her support in the immunohistochemical assays and James Hutchins for correcting the scientific English. The study was supported financially by the ACORN AKC Canine Health Foundation (grant 2118-A).
Highlights
➢ We demonstrated target-specific reference genes to be mandatory for studies in canine reproductive research and medicine.
➢ We defined a universal workflow for normalization of canine endometrial samples.
➢ Two arbitrary normalizers were defined as suitable for tissue- and cell-specific setups.
➢ Definition of arbitrary normalizers reduces the number of necessary reference genes.
➢ Technical variance of LCM replicates was effectively answered with our normalizers.
- Altmae S, Esteban FJ, Stavreus Evers A, Simon C, Giudice L, Lessey BA, et al. Guidelines for the design, analysis and interpretation of 'omics' data: focus on human endometrium. Human reproduction update. 2014; 20:12-28. https://goo.gl/rXvVpz
- Evans GE, Martinez Conejero JA, Phillipson GT, Simon C, McNoe LA, Sykes PH, et al. Gene and protein expression signature of endometrial glandular and stromal compartments during the window of implantation. Fertility and sterility. 2012; 97: 1365-1373. https://goo.gl/hQiU6y
- Evans GE, Phillipson GT, Sin IL, Frampton CM, Kirker JA, Bigby SM, et al. Gene expression confirms a potentially receptive endometrium identified by histology in fertile women. Hum Reprod. 2012; 27: 2747-2755. https://goo.gl/iyUNoQ
- Yanaihara A, Otsuka Y, Iwasaki S, Aida T, Tachikawa T, Irie T, et al. Differences in gene expression in the proliferative human endometrium. Fertility and sterility. 2005; 83: 1206-1215. https://goo.gl/ponvzQ
- Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al: The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin Chem 2013; 59: 892-902. https://goo.gl/KFdfPA
- Bustin SA. Why the need for qPCR publication guidelines?The case for MIQE. Methods. 2010; 50: 217-226. https://goo.gl/ZpVZb6
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009, 55: 611-622. https://goo.gl/J6D6Z6
- Jurcevic S, Olsson B, Klinga-Levan K. Validation of suitable endogenous control genes for quantitative PCR analysis of microRNA gene expression in a rat model of endometrial cancer. Cancer cell international. 2013; 13: 45. https://goo.gl/G3Q77j
- Klein C, Rutllant J, Troedsson MH. Expression stability of putative reference genes in equine endometrial, testicular, and conceptus tissues. BMC research notes. 2011; 4: 120. https://goo.gl/46Demy
- Walker CG, Meier S, Mitchell MD, Roche JR, Littlejohn M. Evaluation of real-time PCR endogenous control genes for analysis of gene expression in bovine endometrium. BMC Mol Biol. 2009; 10: 100. https://goo.gl/icUnbc
- Egenvall A, Hagman R, Bonnett BN, Hedhammar A, Olson P, Lagerstedt AS. Breed risk of pyometra in insured dogs in Sweden. J Vet Intern Med. 2001; 15: 530-538. https://goo.gl/9oLZ4H
- Gibson A, Dean R, Yates D, Stavisky J. A retrospective study of pyometra at five RSPCA hospitals in the UK: 1728 cases from 2006 to 2011. Vet Rec. 2013; 173: 396. https://goo.gl/XdhGLs
- Fransson BA, Lagerstedt A-S, Bergstrom A, Hagman R, Park JS, Chew BP, et al. C-reactive protein, tumor necrosis factor α, and interleukin-6 in dogs with pyometra and SIRS. Journal of Veterinary Emergency and Critical Care. 2007; 17: 373-381. https://goo.gl/nC4UC1
- Hagman R, Ronnberg E, Pejler G: Canine uterine bacterial infection induces upregulation of proteolysis-related genes and downregulation of homeobox and zinc finger factors. PloS one. 2009; 4: 8039. https://goo.gl/bct2a8
- Tamada H, Kawata N, Kawate N, Inaba T, Kida K, Hatoya S, et al. Factors associated with patency of the uterine cervix in bitches with pyometra. Research in veterinary science. 2012; 93: 1203-1210. https://goo.gl/VNVjjw
- Silva E, Henriques S, Brito S, Ferreira-Dias G, Lopes-da-Costa L, Mateus L. Oestrous cycle-related changes in production of Toll-like receptors and prostaglandins in the canine endometrium. J Reprod Immunol. 2012; 96: 45-57. https://goo.gl/1g1DTd
- Silva E, Leitao S, Henriques S, Kowalewski MP, Hoffmann B, Ferreira-Dias G, et al. Gene transcription of TLR2, TLR4, LPS ligands and prostaglandin synthesis enzymes are up-regulated in canine uteri with cystic endometrial hyperplasia-pyometra complex. J Reprod Immunol. 2010; 84: 66-74. https://goo.gl/Pksi36
- Silva E, Leitao S, Ferreira-Dias G, Lopes da Costa L, Mateus L: Prostaglandin synthesis genes are differentially transcripted in normal and pyometra endometria of bitches. Reprod Domest Anim. 2009; 44: 200-203. https://goo.gl/QsJPMB
- Ropka-Molik K, Oczkowicz M, Mucha A, Piorkowska K, Piestrzynska-Kajtoch A. Variability of mRNA abundance of leukemia inhibitory factor gene (LIF) in porcine ovary, oviduct and uterus tissues. Mol Biol Rep. 2012; 39: 7965-7972. https://goo.gl/HRgzSk
- Mulisch M, Welsch U. Romeis Mikroskopische Technik, 18th edition edn. Spektrum Akademischer Verlag Heidelberg. Imprint Springer. 2010. https://goo.gl/3KgvNG
- Jais A, Klein D, Wolfesberger B, Walter I: Gene expression profile of vascular endothelial growth factor (VEGF) and its receptors in various cell types of the canine lymph node using laser capture microdissection (LCM). Veterinary immunology and immunopathology 2011; 140: 207-214. https://goo.gl/27KJn5
- Bartel C, Tichy A, Schoenkypl S, Aurich C, Walter I. Effects of steroid hormones on differentiated glandular epithelial and stromal cells in a three dimensional cell culture model of the canine endometrium. BMC Vet Res. 2013; 9: 86. https://goo.gl/2rsneg
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL: Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012; 13: 134. https://goo.gl/BJSkzh
- Marshall OJ: PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004, 20: 2471-2472. https://goo.gl/m5V5zi
- Zuker M: Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31: 3406-3415. https://goo.gl/wgXy9Y
- Brinkhof B, Spee B, Rothuizen J, Penning LC. Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal Biochem. 2006, 356: 36-43. https://goo.gl/GXZddA
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009; 37: 45. https://goo.gl/smMvjY
- Andersen CL, Jensen JL, Orntoft TF: Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004; 64: 5245-5250. https://goo.gl/Rgvmuc
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP: Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004; 26: 509-515. https://goo.gl/wjr8V3
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002; 3. https://goo.gl/Zdz44R
- Craythorn RG, Girling JE, Hedger MP, Rogers PA, Winnall WR. An RNA spiking method demonstrates that 18S rRNA is regulated by progesterone in the mouse uterus. Mol Hum Reprod. 2009; 15: 757-761. https://goo.gl/w4HqGo
- Bellver J, Mundi M, Esteban FJ, Mosquera S, Horcajadas JA. '-omics' technology and human reproduction: Reproductomics. Expert Review of Obstetrics and Gynecology. 2012, 7: 493-506. https://goo.gl/byFWMj
- Wira CR, Rossoll RM. Antigen-presenting cells in the female reproductive tract: influence of the estrous cycle on antigen presentation by uterine epithelial and stromal cells. Endocrinology. 1995; 136: 4526-4534. https://goo.gl/B5tXgu
- Chen JC, Erikson DW, Piltonen TT, Meyer MR, Barragan F, McIntire RH, et al. Coculturing human endometrial epithelial cells and stromal fibroblasts alters cell-specific gene expression and cytokine production. Fertility and sterility. 2013; 100: 1132-1143. https://goo.gl/gwQuhA
- Wira CR, Grant-Tschudy KS, Crane-Godreau MA. Epithelial cells in the female reproductive tract: a central role as sentinels of immune protection. Am J Reprod Immunol. 2005; 53: 65-76. https://goo.gl/Ktz7Q7
- Ochiel DO, Fahey JV, Ghosh M, Haddad SN, Wira CR: Innate Immunity in the Female Reproductive Tract: Role of Sex Hormones in Regulating Uterine Epithelial Cell Protection Against Pathogens Curr Womens Health Rev. 2008; 4: 102-117. https://goo.gl/sxbDpB
- Fahey JV, Rossoll RM, Wira CR. Sex hormone regulation of anti-bacterial activity in rat uterine secretions and apical release of anti-bacterial factor(s) by uterine epithelial cells in culture. J Steroid Biochem Mol Biol. 2005; 93: 59-66. https://goo.gl/d2eBRp
- Ulbrich SE, Groebner AE, Bauersachs S. Transcriptional profiling to address molecular determinants of endometrial receptivity--lessons from studies in livestock species. Methods. 2013; 59: 108-115. https://goo.gl/xP67tR
- Datta S, Malhotra L, Dickerson R, Chaffee S, Sen CK, Roy S: Laser capture microdissection: Big data from small samples. Histol Histopathol. 2015; 1255-1269. https://goo.gl/XYTzSv
- Spitzer TL, Rojas A, Zelenko Z, Aghajanova L, Erikson DW, Barragan F, Meyer M, et al. Perivascular human endometrial mesenchymal stem cells express pathways relevant to self-renewal, lineage specification, and functional phenotype. Biol Reprod. 2012; 86: 58. https://goo.gl/EF6hbR
- Hines WC, Su Y, Kuhn I, Polyak K, Bissell MJ. Sorting out the FACS: a devil in the details. Cell Rep. 2014; 6: 779-781. https://goo.gl/GDRaJo
- Santin AP, Souza AF, Brum LS, Furlanetto TW. Validation of reference genes for normalizing gene expression in real-time quantitative reverse transcription PCR in human thyroid cells in primary culture treated with progesterone and estradiol. Mol Biotechnol. 2013; 54: 278-282. https://goo.gl/yWiQWW
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005; 6: 279-284. https://goo.gl/MThwzm

Sign up for Article Alerts