Differential Distribution and Evolution of HIV-1 RNA Variants in the Gastrointestinal Tract of Antiretroviral Naïve African AIDS Patients with Diarrhea and (or) Weight Loss?
Phetole Mahasha1*, Edana Cassol1,2, Siva Danaviah3, Robert Bond4, Christopher Seebregts5,6, Schalk W. van der Merwe4,7, Theresa Rossouw1 and Tulio de Oliveira3*
1MRC Unit for Inflammation and Immunity, Department of Immunology, University of Pretoria, South Africa and the Tshwane Academic Division of the National Health Laboratory Service of South Africa
2Department of Health Sciences, Carleton University, Canada
3Africa Centre for Population Studies, University of KwaZulu Natal, Somkhele, South Africa
4Hepatology and GI-Research Laboratory, University of Pretoria, South Africa
5Jembi Health Systems NPC, South Africa
6School of Mathematics, Statistics and Computer Science, University of KwaZulu-Natal, Durban, South Africa
7Department of Internal Medicine, Division of Liver and Biliopancreatic Disorders, University of uven, Belgium
*Address for Correspondence: Phetole Walter Mahasha, MRC Unit for Inflammation and Immunity, Department of Immunology, University of Pretoria, South Africa and the Tshwane Academic Division of the National Health Laboratory Service of South Africa E-mail: thakgalo03@gmail.com
Submitted: 01 December 2015; Approved: 28 December 2015; Published: 03 January 2016
Citation this article: Mahasha P, Cassol E, Danaviah S, Bond R, Seebregts C, et al. Differential Distribution and Evolution of HIV1 RNA Variants in the Gastrointestinal Tract of Antiretroviral Naïve African AIDS Patients with Diarrhea and (or) Weight Loss. SRL Virol Infect Dis. 2016;1(1): 004-016.
Copyright: © 2019 Mahasha P, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Download Fulltext PDF
Background: Obtaining samples of the gastrointestinal tract (GIT) is difficult. As a result, little is known about the diversification and genetic evolution of Human Immunodeficiency Virus-1 (HIV-1) in the GIT. Here, we evaluated the diversity and phylogenetic relationships among HIV-1 RNA quasispecies derived from the duodenum, jejunum, ileum, right colon, left colon and plasma of antiretroviral therapy (ART)-naïve South African AIDS patients infected with HIV-1 subtype C. A total of 45 pol and 350 cloned gag and Env Reverse Transcriptase polymerase Chain Reaction (RT-PCR) fragments were sequenced and subjected to phylogenetic analysis.
Results: Significant differences were observed in the diversity, evolution and trafficking of HIV-1 RNA quasispecies in the small vs. large intestine. Env Ribonucleic acid (RNA) variants were the most diverse with the small intestine exhibiting the highest level of diversity. gag RNA variants exhibited low levels of diversity in all tissues. Migration and exchange of HIV-1 RNA variants between plasma and different regions of the GIT was more evident in gag than in Env trees. Some Env variants formed tight monophyletic clusters of closely related viral quasispecies suggesting there was localized replication and adaptive evolution of the Env gene, primarily in the colon, but also in the small intestine of some patients. Analysis of inferred amino acid sequences indicated that some pol sequences contained substitutions in protein domains associated with altered susceptibility to ART. A small number of differentially distributed Env variants had substitutions that are known to alter the neutralization and co-receptor binding properties of the V3 loop.
Conclusions: Diversification of HIV-1 in the GIT of ART-naïve AIDS patients is a highly dynamic process involving localized, tissue-specific evolution in addition to extensive exchange of HIV-1 RNA variants between plasma and different gut compartments.
KEYWORDS
Gastrointestinal tract (GIT); HIV-1; RNA quasispecies; Genetic diversity
ABBREVIATIONS
ART: AntiRetroviral Therapy; GALT: Gut Associated Lymphoid Tissue; GIT: Gastrointestinal Tract; HAART: Highly Active Anti Retroviral Therapy
INTRODUCTION
The gastrointestinal tract (GIT) is a complex organ composed of multiple tissue compartments with different structural and functional elements and diverse cell types. Due to its continuous exposure to a wide range of microbes and food antigens, the GIT is in a constant state of “physiological” inflammation and contains a large number of activated CCR5+ memory CD4+ T cells, the primary target of HIV-1. As a result, the GIT is a site of intense viral replication and severe CD4+ T cell depletion (Veazey et al., 1998; Guadalupe et al., 2003; Mehandru et al., 2004; Brenchley et al., 2004; Mattapallil et al., 2005; Guadalupe et al., 2006; Centlivre et al., 2007), beginning during primary infection (Guadalupe et al., 2003; Mehandru et al., 2004; Centlivre et al., 2007) and continuing at a lesser pace throughout the chronic phase of infection (Brenchley et al., 2004). This is accompanied by progressive immunological and structural damage to the gut associated lymphoid tissue (GALT), a loss of epithelial integrity, increased microbial translocation and both local and systemic immune activation (Brenchley et al., 2006; Paiardini et al., 2008; Brenchley et al., 2008). Antiretroviral therapy (ART) is only partially effective in restoring the immunological and structural integrity of the intestinal mucosa (Mehandru et al., 2006; poles et al., 2006; Cassol et al., 2010), even when initiated during primary infection (Mehandru et al., 2006).
Despite the importance of the GIT in the pathogenesis of HIV-1 infection, and as a reservoir of treatment refractory virus, little is known about the diversity and genetic evolution of HIV-1 variants in different compartments of the intestine, or whether these diverse variants are associated with differing pathologies. There is also a paucity of information relating to the dissemination and genetic relationships between intestinal variants and those found in other tissues including the circulation. This is due, in large measure, to difficulties and ethical considerations inherent in sampling the GIT and to the small size of the samples obtained. Most studies have been confined to one or two anatomical sites (poles et al., 2001; Avettand-Fenoel et al., 2011; Imamichi et al., 2011; Lerner et al., 2011; Evering et al., 2012) and have involved the sequencing of proviral DNA isolated from patients on suppressive ART (Lerner et al., 2011; Evering et al., 2012). DNA contains archived and defective genetic material and may not accurately reflect the full spectrum of replication competent viral strains while studies of patients on highly active ART (HAART) with low levels of viral replication may underestimate HIV-1 diversification. A study by Avettand-Fenoel et al., found that prior to the introduction of HAART, HIV-1 DNA variants in the rectum were more diverse than those in blood. However, in patients on HAART, there was an “equalization” of viral dynamics in the rectum and blood. As a result, in HAART-treated patients, there was no difference in the degree of viral diversity between the rectum and peripheral blood (Avettand-Fenoel et al., 2011).
We recently reported that there are marked differences, not only in the level of CD4+ T cell depletion and viral replication, but also in immune activation and ART-induced CD4+ T cell restoration in the small versus large intestine of South African AIDS patients with chronic diarrhea and (or) unexplained weight loss (Cassol et al., 2013). The aim of the current study was to determine whether these different intestinal Environments have an impact on HIV-1 genetic diversification. To address this issue, we assessed the diversity and investigated the phylogenetic relationships among HIV-1 RNA variants in the plasma, duodenum, jejunum, ileum, right and left colon of a subset of our ART-naïve ADIS cohort. An understanding of HIV-1 diversification in different regions of the GIT is important, not only for the design of strategies aimed at interrupting disease progression, but also for the optimization of ART and the assessment of therapeutic efficacy.
MATERIALS AND METHODS
Patients: Ten ART-naive patients with a complete GIT sample set were selected for genetic studies. Three patients, HIV019, HIV021 and HIV029, who had sufficient sample were selected for extensive phylogenetic analysis of HIV-1 RNA quasispecies. These patients were recruited at the Comprehensive Care, Management and Treatment Clinic in Pretoria as part of a previously described study of ART-induced CD4+ T cell restoration in South African patients with advanced HIV-1 infection (WHO stage III or IV), chronic diarrhea (duration of >4 weeks) and (or) unintentional weight loss (>10% from baseline) of unknown etiology. Patients were eligible for study if their sputum tested negative for Mycobacterium tuberculosis and their stool samples were culture negative and stained negative for fungal and parasitic organisms (Cryptosporidium, Microsporidia and Isospora) as determined by hematoxylin-eosin, periodic acid-Schiff diastase, Ziehl-Neelsen and Giemsa staining. The study protocol was reviewed and approved by the Faculty of Health Sciences Research Ethics Committee of the University of Pretoria and written informed consent was obtained from all study participants.
Specimen Collection
“Pinch biopsies” were collected from 5 different sites in the small and large intestine. The duodenum and jejunum were sampled using a Fujinon double balloon enteroscope and Radial III biopsy forceps (Boston Scientific). Endoscopic biopsies of the terminal ileum, right and left colon was obtained using a standard colonoscope. Biopsies were placed on ice in RPMI 1640/10% fetal calf serum (FCS) and immediately processed for flow cytometry or snap frozen in liquid nitrogen and stored at -80oC for viral load (VL) and sequence analysis.
HIV-1 RNA Quantification, cDNA synthesis and PCR Amplification
RNA was extracted from plasma and biopsy samples using the NucliSENS easyMAG extraction kit from BioMerieux and quantified using the Nuclisens Easy HIV-1 QT v.1.2 kit. Results were expressed as the number of HIV-1 RNA copies per gm of tissue or mL of plasma. RNA extracted from gut biopsy samples was reverse transcribed using Superscript III and RNaseOUT (Invitrogen, San Diego, CA) and oligodT or KVL065 (Van Laethem et al., 2006) as primers for the synthesis of Env and gag cDNA, respectively. Reverse transcription of the pol gene was performed using the MMLV Reverse Transcriptase 1st Strand cDNA Synthesis Kit (Epicentre Biotechnologies, Madison, Wisconsin) and the reverse primer, IN3 (Pillay, 2006). Following digestion with RNase H, cDNAs were purified using the Invitrogen (Env) or the GeneJET RNA (gag, pol) Purification Kits (Fermentas Life Sciences, Burlington, Ontario). Nested PCR of pol (1856), Env (667 bp) and gag (1997 bp) encoding regions was performed on 0.2 µg of cDNA using either Platinum Taq High Fidelity DNA polymerase (Invitrogen, San Diego, CA) or Excel Taq DNA polymerase (ExSel, JMR Holdings Inc., London, UK) and the following primer sets: MK605/CD4R2 and ES7/ES8 for Env (Gordon et al., 2003); KVL064/KVL065 and KVL66/KVL67 for gag (Van Laethem et al., 2006); and G25Rev/IN3 and AV150/polM4 for pol (Pillay, 2006). RT-PCR products were purified using the GeneJET™ Gel Extraction (Fermentas Life Sciences) or Purelink PCR Purification (Invitrogen) kits. pol RT PCR products were sequenced directly using published primers: AV150, polM4, SQ5F, SQ12R, SQ6F, and 13R(2)C(2) (Pillay, 2006; Rousseau et al., 2006).
Cloning and Sequencing
gag and Env RT-PCR products were cloned into the pCU®4 TOPO TA vector (Invitrogen) and transformed into XL 10-Gold Ultra-competent cells (Agilent Technologies, Santa Clara, CA). Plasmids containing the expected inserts, as confirmed by restriction digestion with FastDigest EcoRI (Fermentas Life Sciences), were purified using the Wizard® SV 96 Plasmid DNA Purification System (Promega Corporation, Madison, WI). Plasmid DNA was quantified and sequenced on an automated ABI 3100 DNA analyzer using the Big-Dye terminator v3.1 sequencing kit (ABI, Foster City, CA) and the following primers: T7 and M13R-ES8 for Env (Gordon et al., 2003) and KVL066, KVL067, KVL078, KVL079, KVL080, GA1, AV103 and AV159 for gag (Van Laethem et al., 2006).
Sequence Alignment, Editing and Subtype Analysis
Chromatograms were imported and edited in CLC DNA workbench 5 () or Geneious version 5.3.6 (http://www.geneious.com. To rule out contamination, each new sequence was compared to other sequences amplified at the same time and to HIV-1 sequences previously amplified in our laboratory. Sequences were aligned using CLUSTAL W and manually edited using Bioedit version 7.0.4.1 (Thompson et al., 1994) and/or Se-AL (http:tree.bio.ed.ac.uk). Gap-stripped sequences were subtyped using the Rega version 3.0 (http://www.bioafrica.net/rega-genotype/html/subtypinghiv.html) and COMET (http://comet.retrovirology.lu/index.php) subtyping tools and compared to other subtype C strains from South Africa (Los Alamos subtype database, ).
Genetic diversity
Pairwise distances among RNA quasispecies in the plasma, duodenum, ileum, jejunum, right and left colon were calculated using the Kimura 2 parametric model of nucleotide substitution implemented in MEGA (Kimura, 1980; Tamura et al., 2011). The gamma distribution model was used to estimate the rate of variation among sites and a bootstrap of 1000 replications was used to calculate the mean distance and standard error. The means of all pairwise genetic distances for each quasispecies relative to other Env or gag quasispecies within and between different tissue compartments was also calculated.
Phylogenetic analyses
Nucleic acid sequences were subjected to extensive phylogenetic analysis. Trees were constructed using Maximum Likelihood (ML) (Guindon et al., 2010) and Bayesian tree inference methods (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) in PhyML and BEAST software packages, respectively. Both methods were performed using the Gamma heterogeneity alpha parameter. The SPRs method and a BioNJ starting tree implemented in PhyML were used to optimize and confirm ML tree topology. The reliability of branching orders was confirmed by performing 1,000 bootstrapping re-samplings. Values >70% were considered significant. The reproducibility of Bayesian trees was assessed using four MCMC chains of 10,000,000 generations sampled every 1,000 generations. Results were visualized in Tracer (Drummond and Rambaut, 2011) and the first 10% of sampled trees and their estimated parameters were excluded from the final analysis (i.e. burning). All the parameters had effective sample sizes greater than 300, suggesting good mixing and sampling of trees. BEAST version 1.6 (Drummond et al., 2012) was used to investigate migration and estimate the most recent common ancestor (MRCA). The initial tree was generated using a simple, constant population size coalescent model. The rate of nucleotide substitution was estimated using a general time reversible (GTR) substitution model with gamma distribution. An uncorrected relaxed clock with gamma distribution and an initial chain of 108 was used to determine the time of divergence of each compartment. A burning of 10% was applied with the initial 10% of data points being excluded from the analysis. The log file output was viewed in Tracer and was deemed suitable for analysis if the estimated sample sizes were greater than 100. Posterior probability support for the evaluation of the internal branches was estimated using FIGTREE. TreeAnnotator [] was used to generate and visualize the maximum clade credibility (mcc) trees.
Amino acid sequence analysis
Nucleic acid sequences were translated, aligned and used to obtain an inferred amino acid sequence for each of the 350 cloned HIV-1 quasispecies amplified from the plasma, duodenum, jejunum, ileum and colon of patients HIV019, HIV021 and HIV029. Each amino acid sequence was screened for integrity of the coding sequence (deletions, insertions, stop codons) and compared to the consensus sequence for that tissue compartment and to the subtype C consensus sequence for southern Africa. Three different methods were used to predict the tropism and co-receptor usage (CCR5 vs CXCR4) of the V3 loop: the Web Position Specific Scoring Matrix (PSSM) (Jensen et al., 2003), the geno2pheno (Lengauer et al., 2007) and the CoRSeqv3C (Cashin et al., 2013) prediction algorithms. pol amino acid sequences were screened for substitutions at positions associated with primary and accessory drug resistance to nucleoside reverse transcriptase (RT) and non-nucleoside RT inhibitors (NRTIs, NNRTIs) and protease inhibitors (PIs) as determined using the HIV Drug Resistance Database from Stanford University site (/hiv/).
Nucleotide sequence accession numbers
Genbank accession numbers for the sequences obtained in this study are: KF905653- KF905865, for Env gene sequences; KF905866- KF906024, for gag gene sequences; KF906025- KF906069, for pol gene sequences.
RESULTS
Patient
All ten patients were ART-naïve and had advanced HIV-1 infection (WHO stage III or IV). Symptoms leading to endoscopy were diarrhea in 8 and unexplained weight loss of >10% in 2 patients. The mean peripheral blood CD4+ T cell count (± SD) at the time of presentation was 102 ± 65 cells/µL; the mean plasma VL (± SD) was 5.41 ± 5.57 log10 copies of HIV-1 RNA/mL and the mean age (± SD) was 37 ± 10.6 years. The mean % of CD4+ T cells ( SD) in the GIT ranged from 3.2 (± 2.28) in the duodenum to 3.8 (± 4.28) in the jejunum, 8.9 (±9.92) in the ileum and 9.8 (± 7.66) in the right colon. VLs ranged from 3.68 (± 1.32) log10 HIV-1 RNA copies/gm of tissue in the duodenum to 3.30 (± 1.29) in the jejunum, 4.27 (± 0.80) in the ileum and 4.39 (± 1.31) in the right colon. Non-specific mild-to-moderate enteritis or enterocolitis was detected in at least one site in 70% of patients, at a similar frequency in all four intestinal sites (70%, 70%, 80% and 70% in the duodenum, jejunum, ileum and colon, respectively).
Genetic diversity
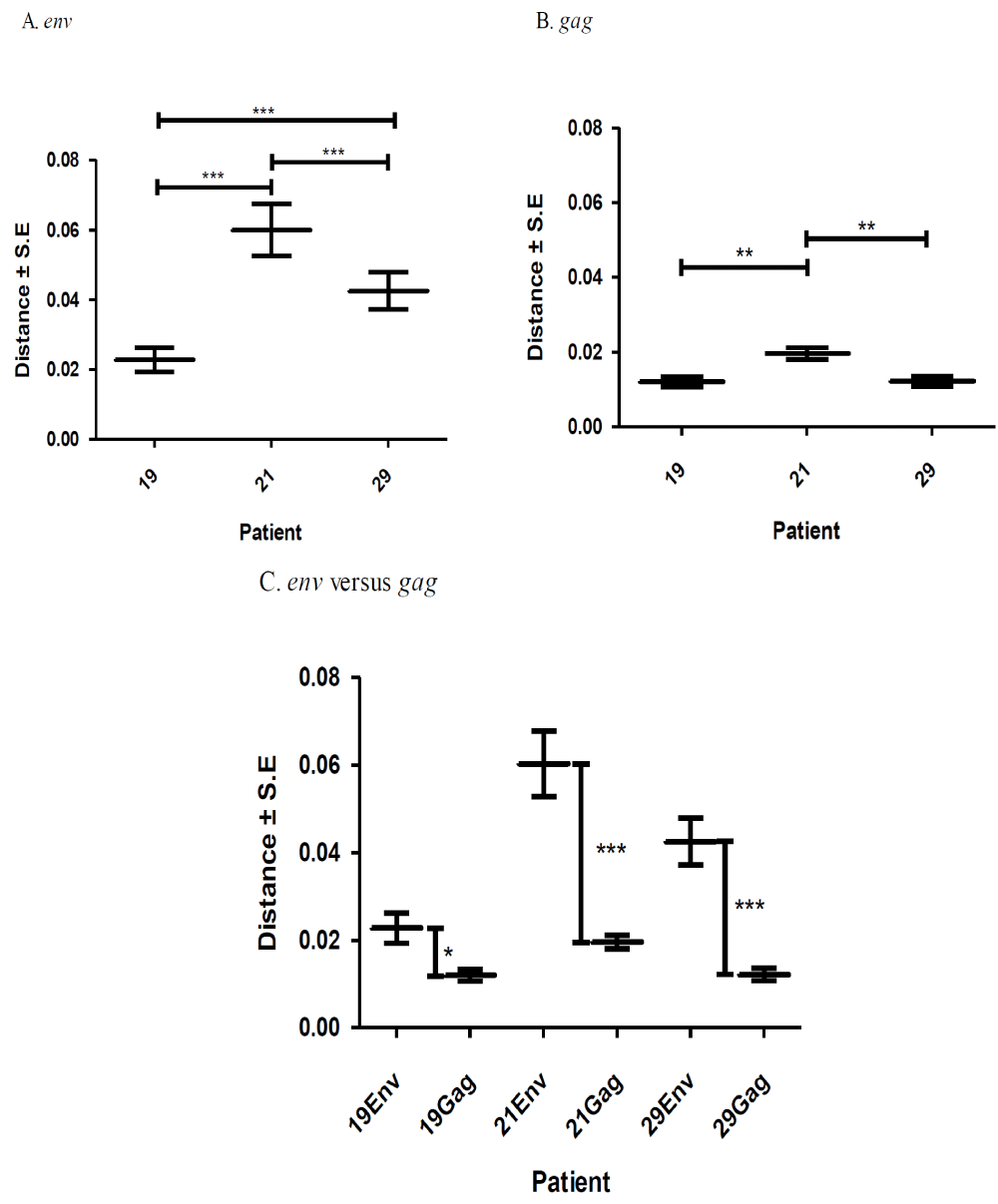
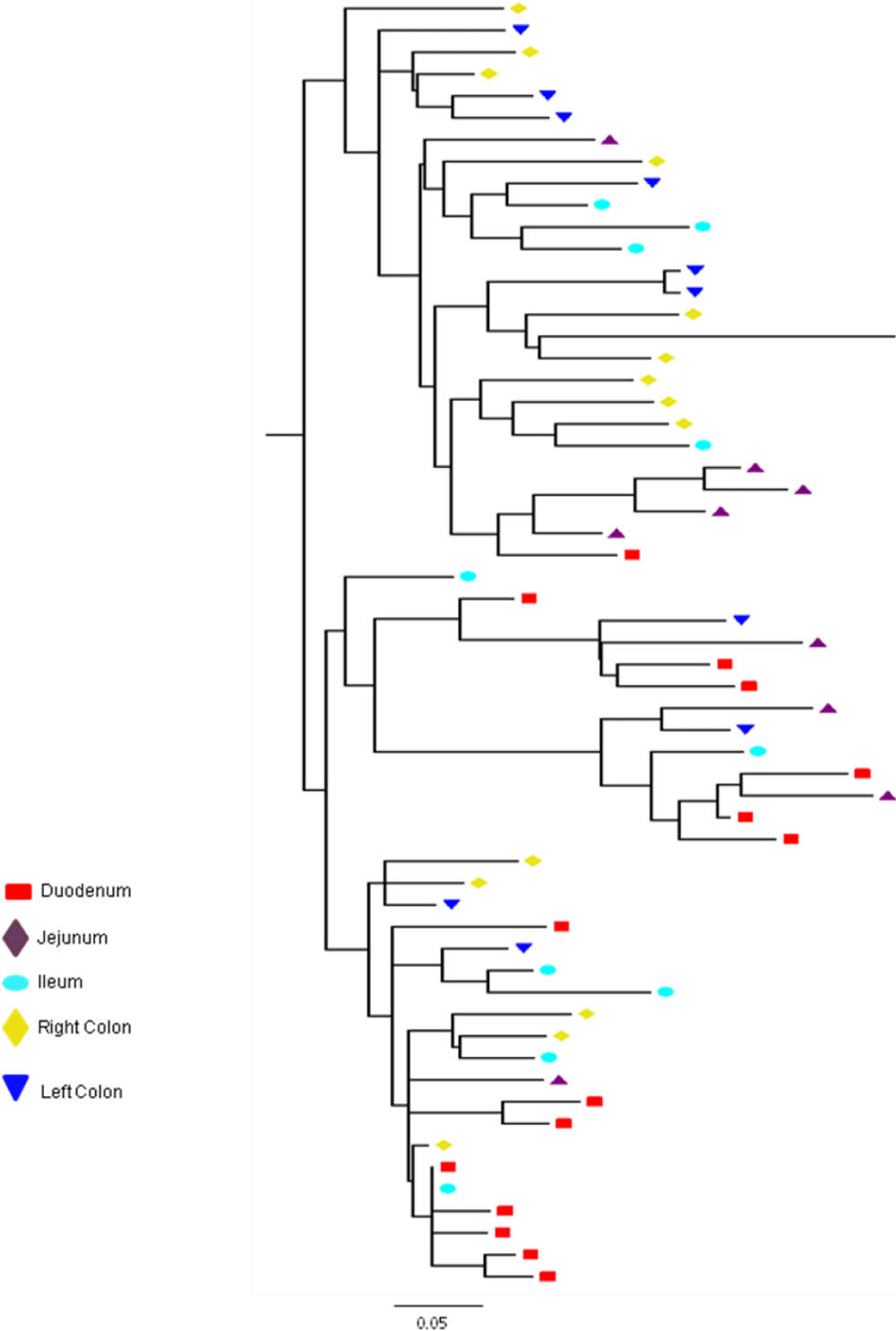
A total of 395 sequences [45 pol RT PCR and 350 cloned Env and gag HIV-1 RNA quasispecies obtained from the duodenum (n = 61), jejunum (n = 72), ileum (n = 73), right colon (n = 80) and left colon (n = 64) of patients HIV019, HIV021 and HIV029 were analyzed. All of these sequences were identified as subtype C when analyzed by REGA and COMET (bootstrap values of >95%). Diversity divergence, estimated in Mega [28, 29]. differed significantly among different patients and between the Env and gag gene regions. Mean overall pairwise distances among Env HIV-1 RNA variants ranged from 2.27% (S.E. 0.35%) in patient HIV019 to 4.25% (S.E. 0.54%) in patient HIV-29 and 5.99% (S.E. 0.74%) in patient HIV021 [p<0.001]. Overall pairwise distances among gag HIV-1 RNA variants ranged from 1.20% (S.E., 0.14%) in patients HIV019 to 1.22% (S.E., 0.14%) in patient HIV029 and 1.96% (S.E., 0.16%) in patient HIV021 [p<0.01]. For each patient, the diversity among Env RNA quasispecies was significantly greater than among gag RNA quasispecies (p<0.05 for patient HIV019; p<0.001 for patients HIV021 and HIV029) Figure 1.
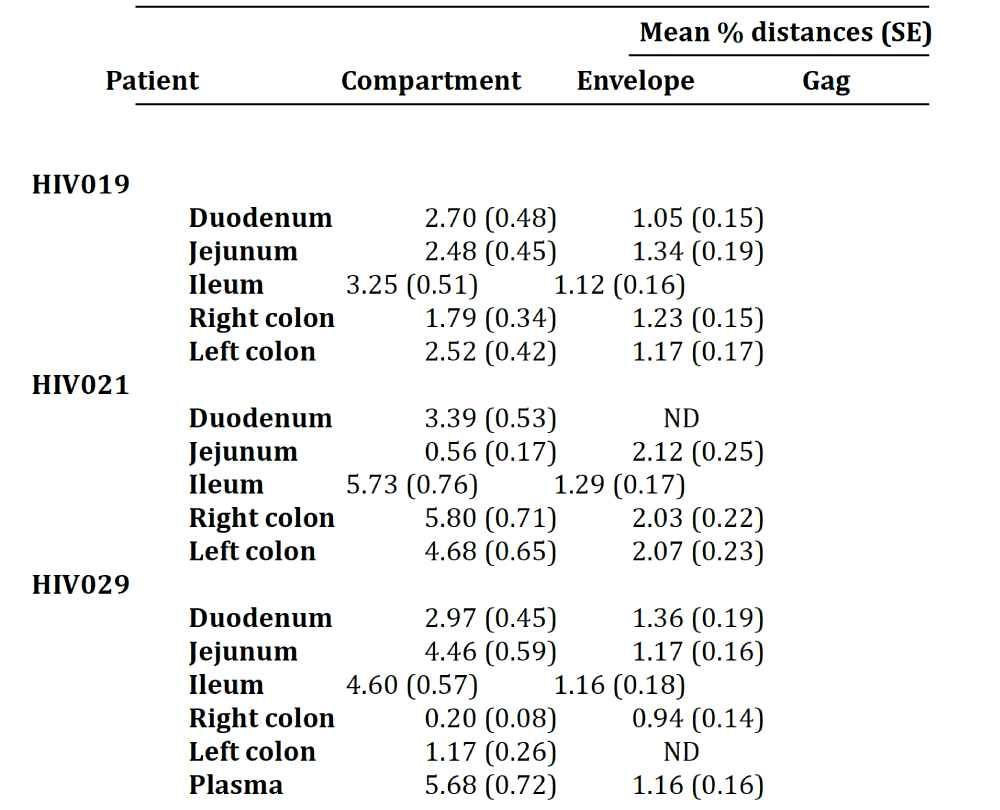
Next, we calculated the pairwise distances for each individual clone obtained from patients HIV019 (72 Env, 59 gag), HIV021 (66 Env, 44 gag) and HIV029 (65 Env, 44 gag) compared to every other cloned quasispecies obtained from the same tissue of that particular patient. The number of clones analyzed for each individual compartment is shown in Table 1. In general, Env variants in the small intestine were more diverse than those in the colon Figure 2 and with few exceptions, most notably the jejunum of patient HIV021 and the right and left colon of patient HIV029, the diversity of Env was greater than that of gag variants in the same tissue. Depending on the patient, inter-compartmental distances ranged from 0.20% to 5.80% among Env and from 0.94% to 2.12% among gag HIV-1 RNA variants Table 2. As described below, the lower than expected diversity among Env sequences in the jejunum of patient HIV021 and the colon of patient HIV029 was due to the presence of large monophyletic clusters of closely related quasispecies.
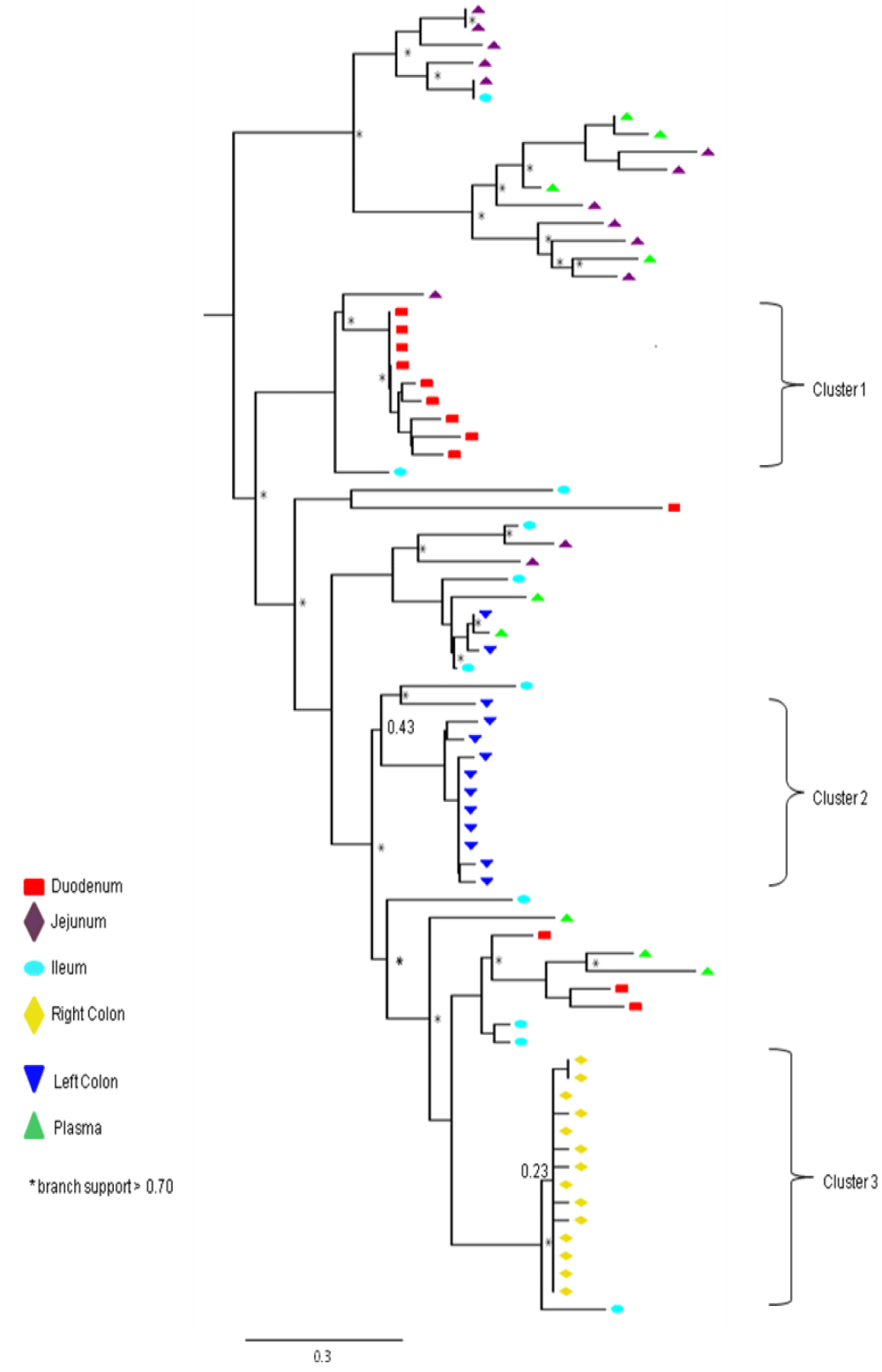
Phylogenetic analysis of pol RT PCR products reveals extensive heterogeneity in the consensus sequences derived from different regions of the GIT
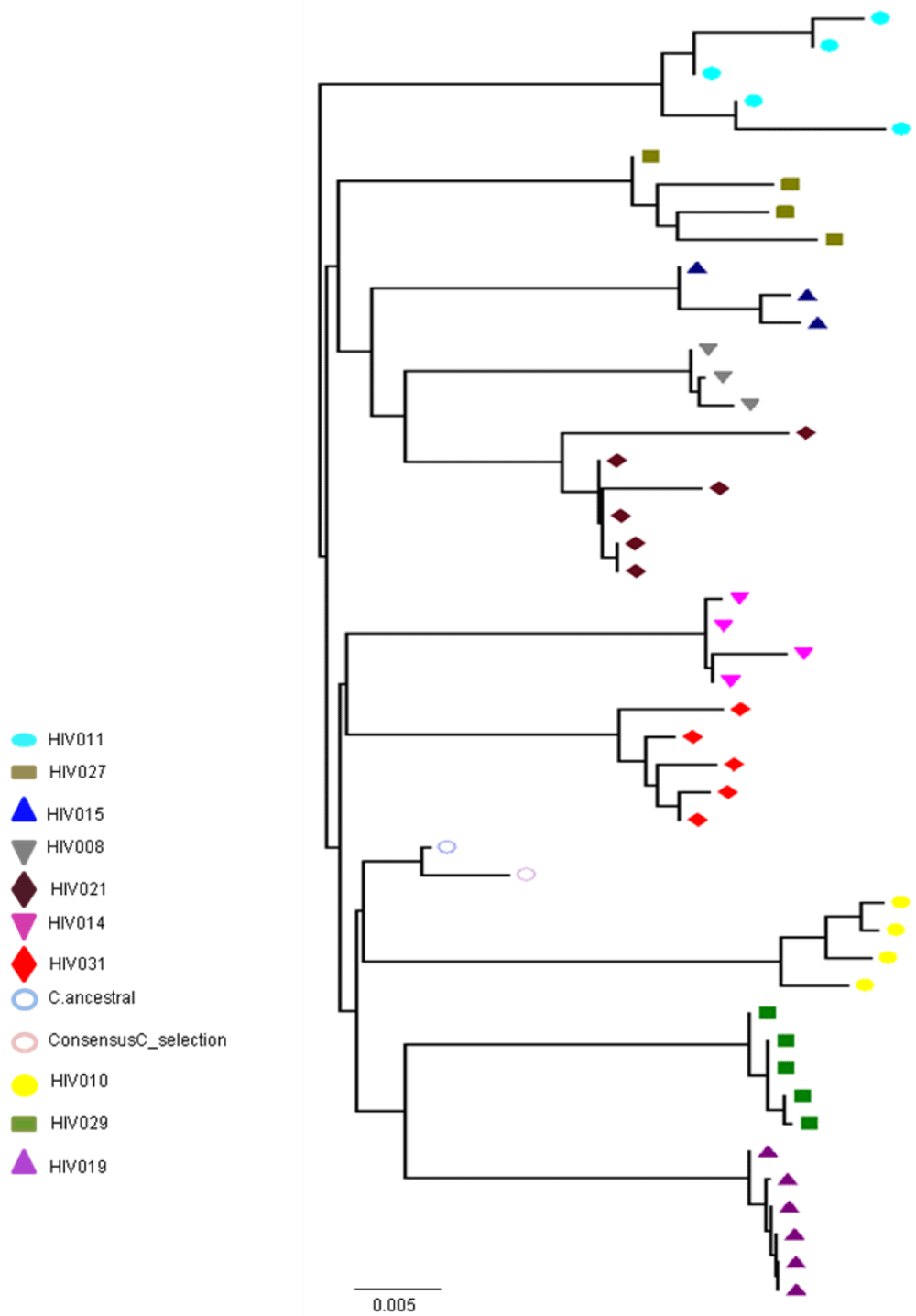
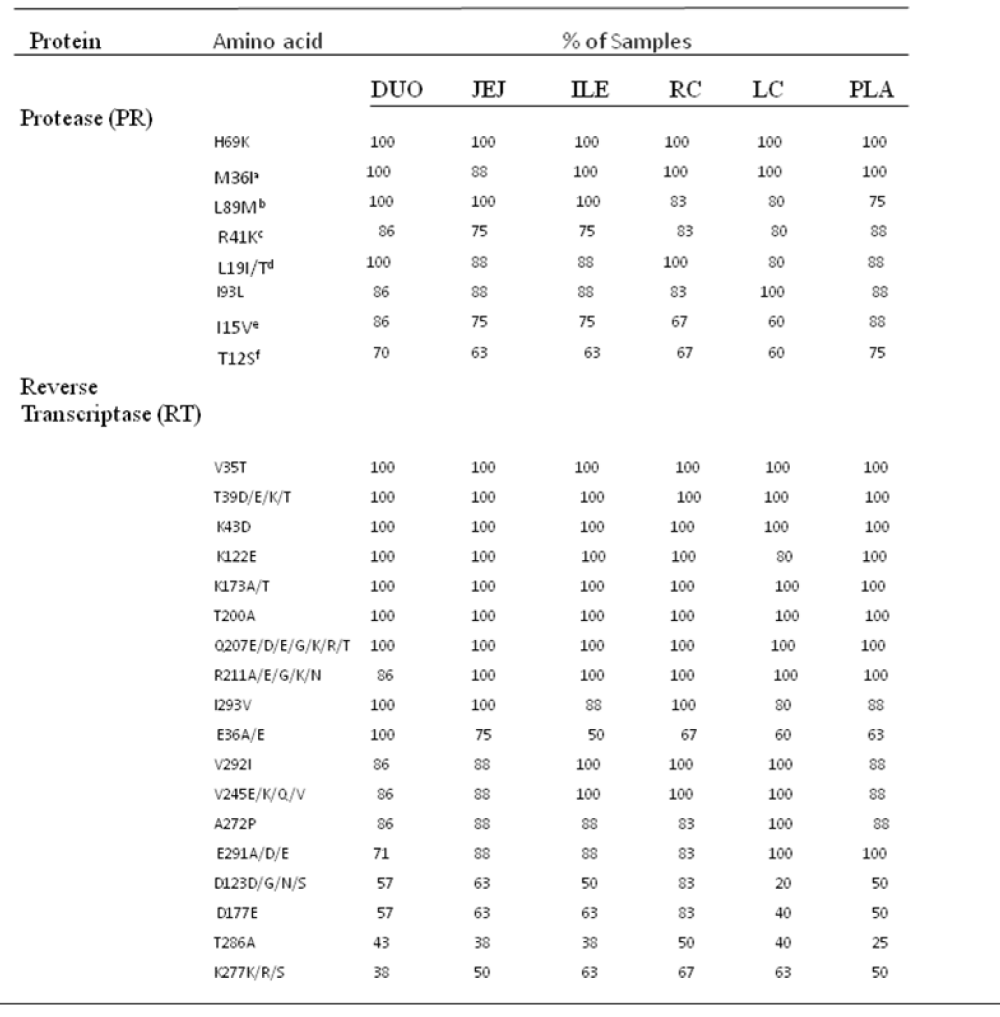
As shown in Figure 3, the nucleotide sequences obtained from all 45 pol RT PCR products were unique and clustered by individual patient and tissue compartment with no cross-contamination of sequences (bootstrap values >70%). This pattern was observed irrespective of whether ML or Bayesian methods were used for tree construction. As expected, no primary drug resistance mutations were detected in the GIT, or in the plasma of the ten patients studied. However, at the amino acid level, we did detect a number of differentially distributed polymorphisms at positions that are known to cause resistance to RT inhibitors. One substitution, RT D67G, was detected in the colon, but not in the plasma or small intestine of patient HIV010. A second highly unusual substitution, RT L210V, was detected in the ileum of patient HIV011. Although D67N and L210W are well-known resistance mutations, the impact of D67N and L210V substitutions on drug susceptibility are not known. The mean number of substitutions in the RT gene relative to HXB2 was 23.2 2.3 (range 18-37) with 86.0% of sequences having 21 or more substitutions relative to HXB2. Some substitutions, such as D123D/G/N/S and T286A, exhibited extensive inter-patient and inter-compartmental variation while others, including V35T, V245Q, T200A and I293V, were highly conserved among different patients and different tissues of the GIT Table 3. polymorphisms associated with altered susceptibility to protease inhibitors were also differentially distributed. Two of these substitutions, T74S and V77I, were detected in the plasma, duodenum, jejunum, ileum and right colon of patient HIV011. PR T74S is present in up to 10% of ART-naive patients infected with HIV-1 subtype C and is associated with decreased susceptibility to nelfinavir (NFV). PR V77I, is a common to compensatory mutation that is selected by NFV and saquinavir (SQV). Other protease polymorphisms that are common in subtype C viruses, M36I/R41I/ H69K/L89M and T12S/I15V/L19I were present in most, but not all, patients and compartments of the GIT Table 3. The first series of substitutions is associated with increased catalytic activity of the protease enzyme (Velazquez-Campoy et al., 2001); the second series, when present in combination with I93L, has been linked to increased sensitivity to lopinavir (Gonzalez et al., 2003).
Phylogenetic analysis of HIV-1 RNA quasispecies reveals extensive trafficking and mixing of variants among different regions of the GIT and between the GIT and plasma
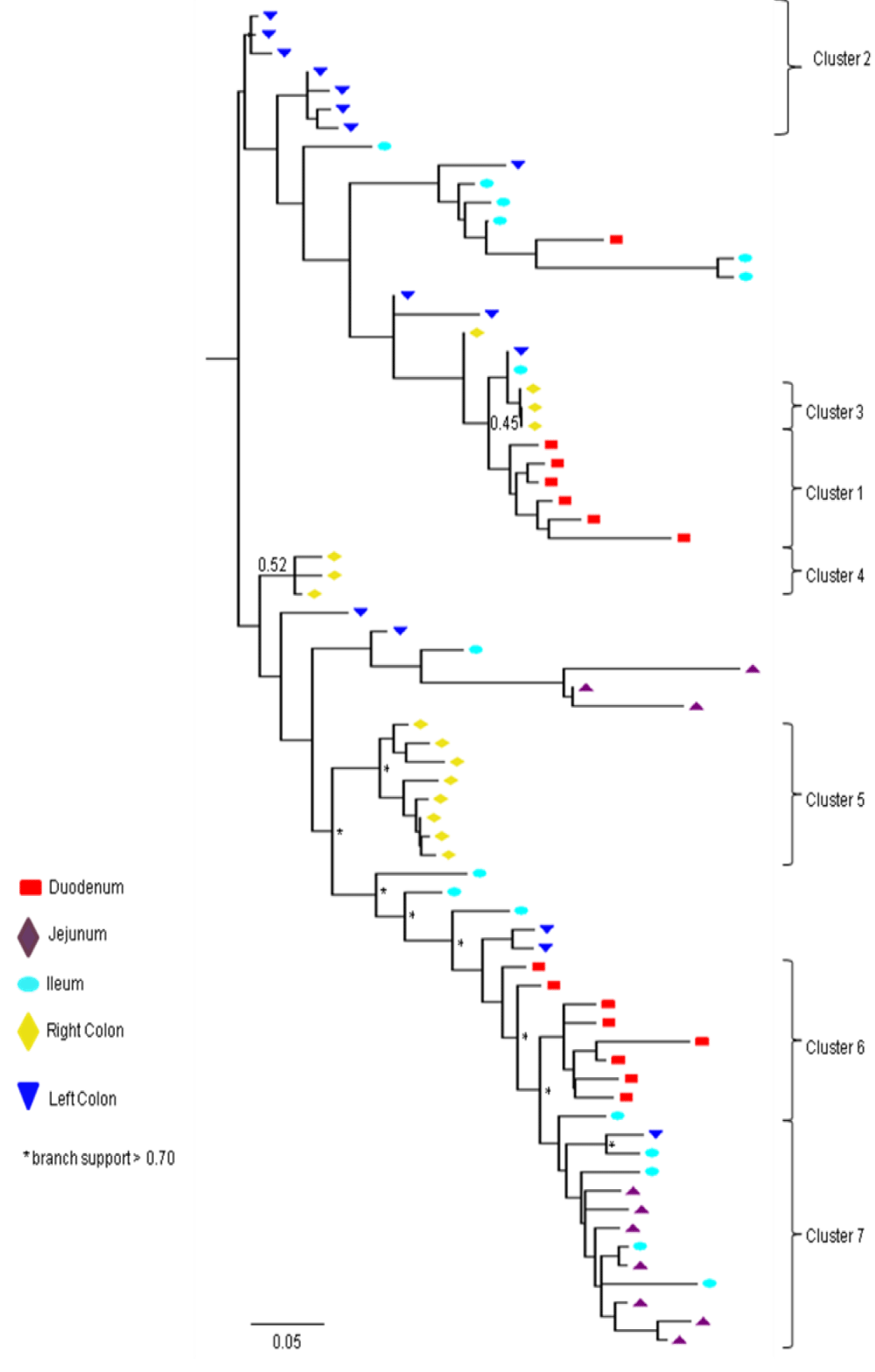
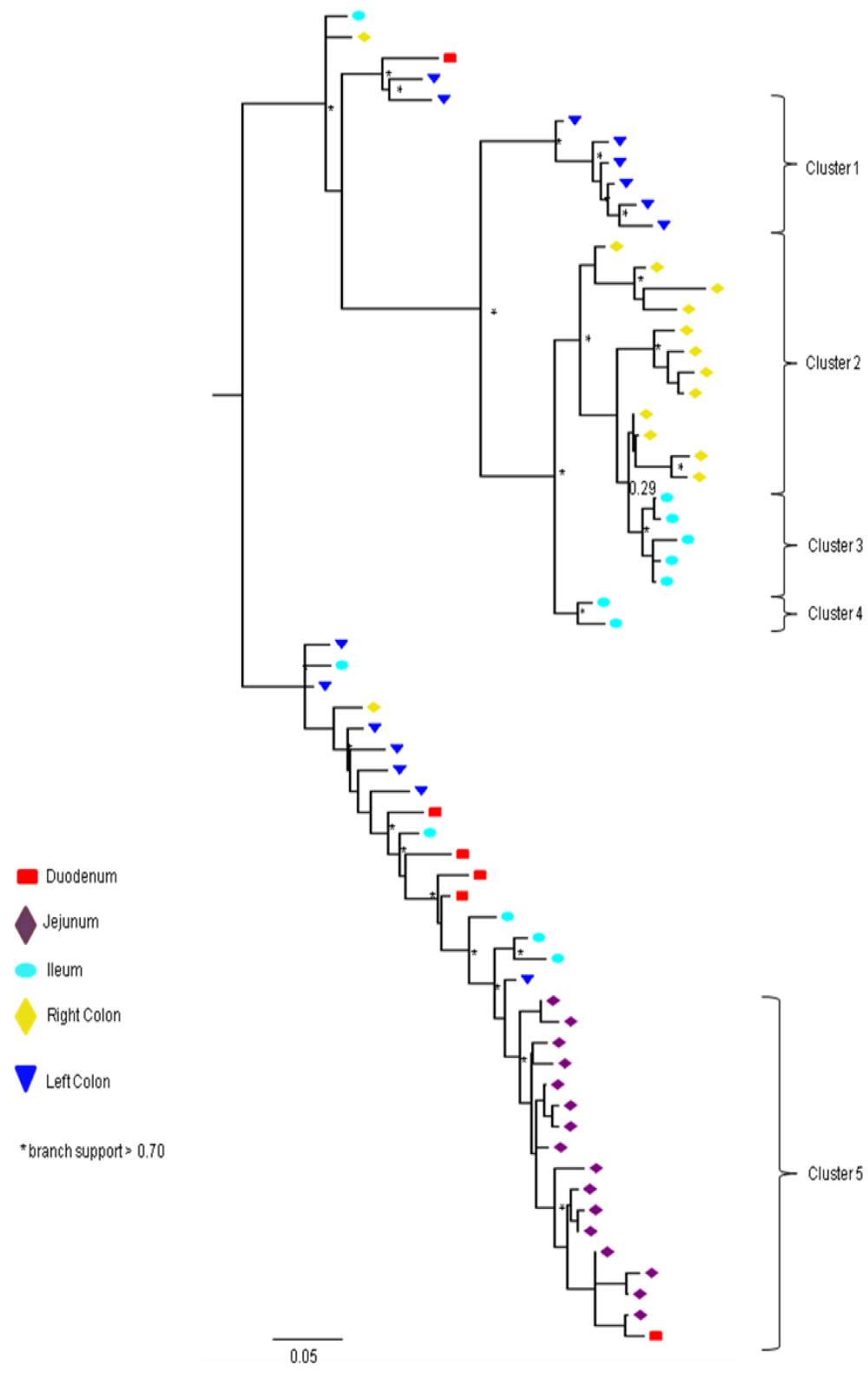
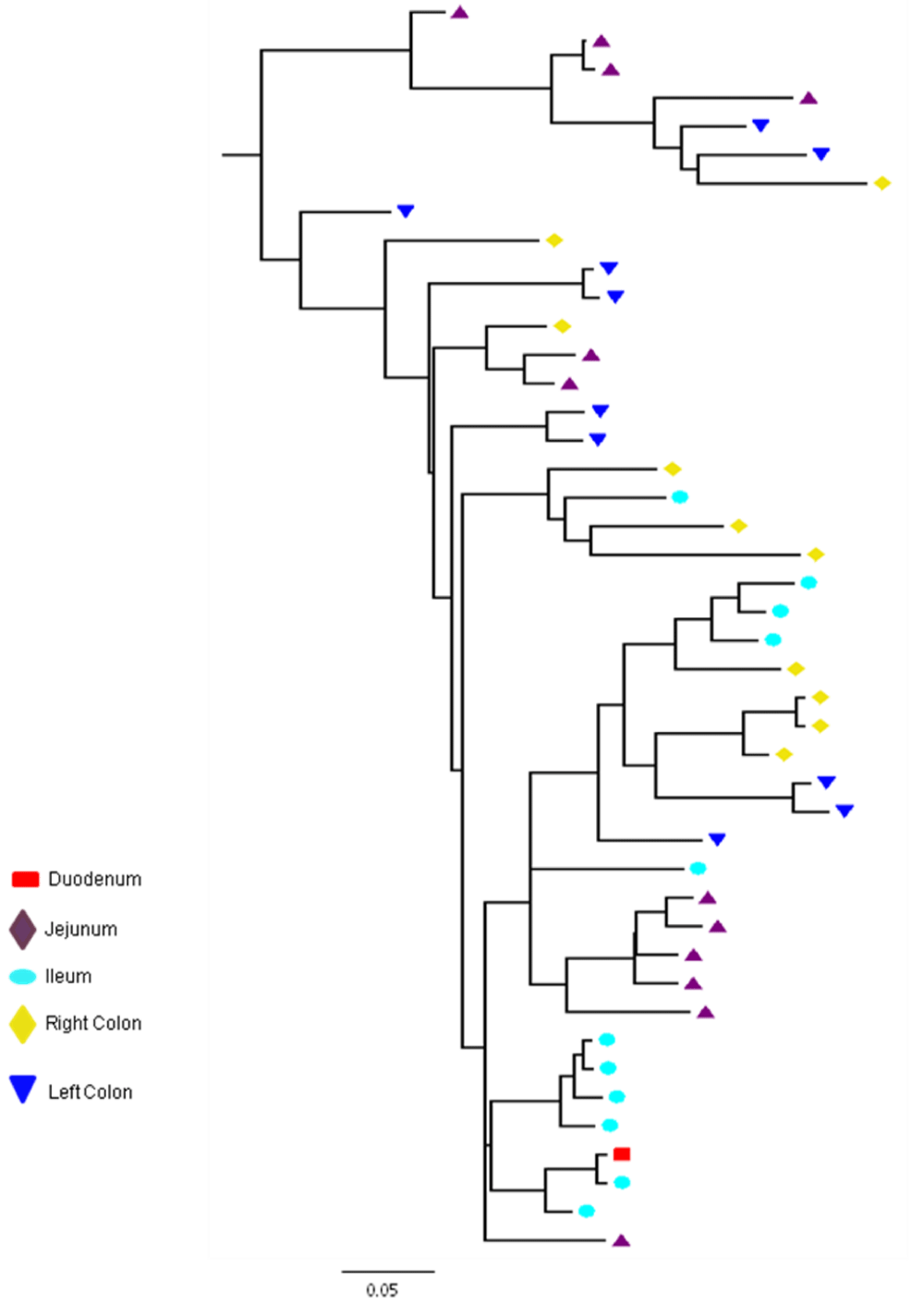
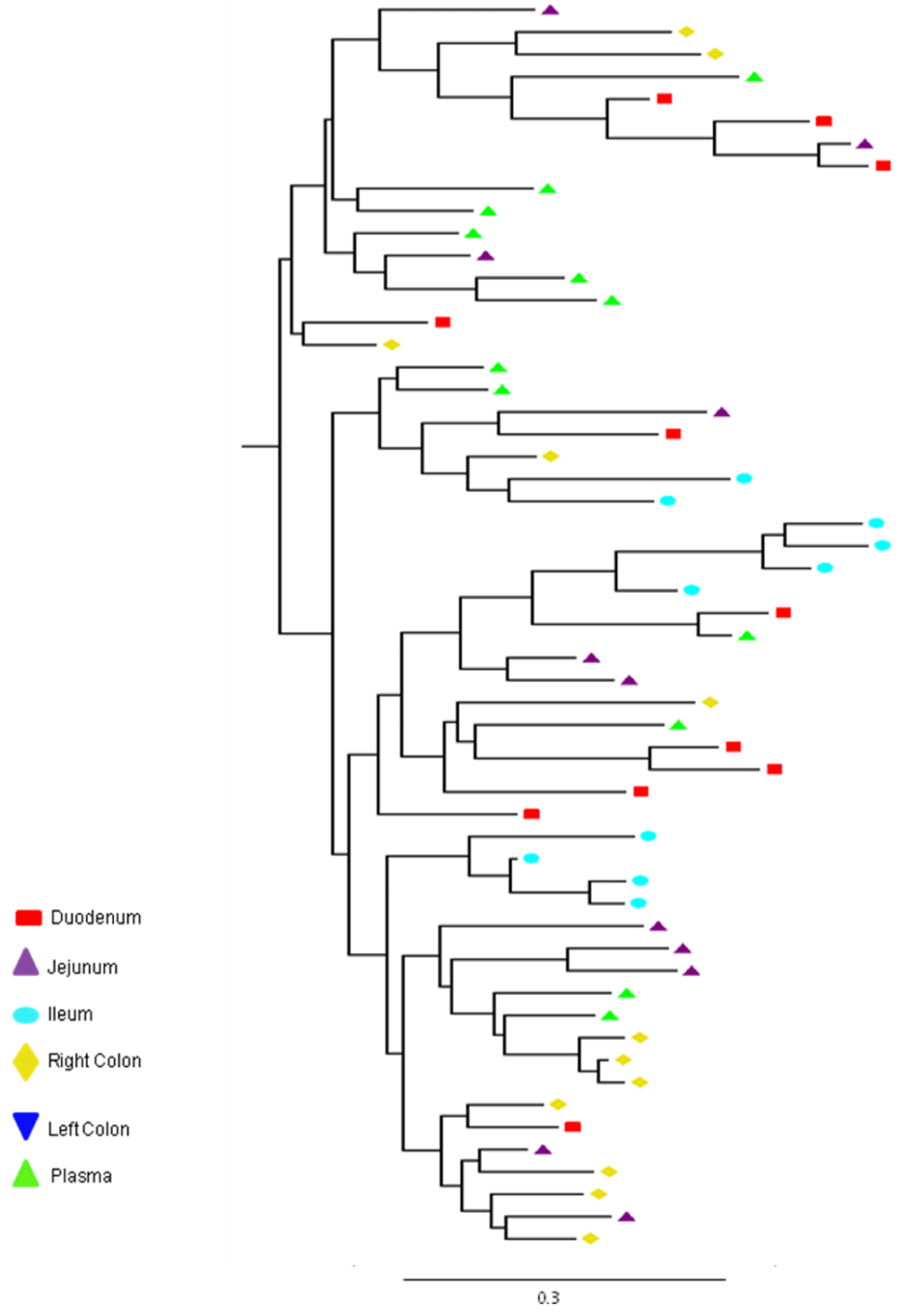
Partial or total inter-compartmental mixing of RNA quasispecies was observed among different compartments in the GIT of patients HIV019, HIV021 and HIV029 Figure 4, A-F. Mixing of HIV-1 RNA was more evident for the gag than the Env gene with extensive exchange of gag quasispecies being observed among all 5 sites in the small and large intestine, and between the GIT and plasma of patient HIV029 Figure 4, D-F. Mixing of Env HIV-1 RNA was less pronounced and observed primarily in the small intestine (duodenum, jejunum and ileum). In addition to mixed clusters, distinct monophyletic clusters of closely related Env HIV-1 RNA variants were detected in the right and left colon of all 3 patients Figure 4, A-C and in the duodenum of patient HIV019, and the jejunum and ileum of patient HIV021. The presence of these tight clusters suggests there was localized tissue-specific evolution of the HIV-1 Env gene, especially in the right and left colon, and in the small intestine of some patients. In contrast, distinct clustering of gag RNA quasispecies was not observed Figure 4, D-F suggesting that trafficking, rather than virus evolution, was the main factor contributing to the heterogeneity of the gag encoding gene region in the GIT of AIDS patients with diarrhea and/or weight loss.
Clustering of Env RNA quasispecies is most consistent in the colon
Clustering was most evident in patient HIV029 where 69% (9/13) of Env HIV-1 RNA variants in the duodenum, 85% (11/13) of variants in the left colon and 100% (14/14) of variants in the right colon segregated as single monophyletic clusters (Figure 4C, clusters 1, 2 and 3, respectively) with posterior probability scores of 1.0, 0.48 and 0.35, respectively. Similarly, 93% (14/15) of Env HIV-1 RNA sequences in the right colon of patient HIV019 fell into 3 distinct clusters (Figure 4A, clusters 3, 4 and 5) with posterior probabilities ranging from 0.47 to 0.99. A notable feature of patient HIV021 Figure 4B, was the relationship between Env HIV-1 RNA sequences in the colon and the ileum. These sequences were found in 4 clusters that segregated together on a single branch of the tree: a basal cluster consisting of 6 sequences from the left colon (cluster 1); an external cluster (cluster 2) containing the majority (12/14, 86%) of Env sequences from the right colon and two closely linked clusters of Env RNA from the ileum (clusters 3 and 4), one of which was a mixed ileal/colonic cluster (cluster 4). The posterior probability for these clusters was 1.0, 1.0, 0.93 and 0.96, respectively. In contrast to the lower GIT, most Env HIV-1 RNA variants in the small intestine were inter-dispersed among the duodenum, jejunum and ileum. Exceptions included two Env RNA clusters in the duodenum of patient HIV019 (clusters 1 and 6) and a large cluster of 17 closely related Env RNA sequences in patients HIV021, 16 from the jejunum and 1 from the duodenum. Many sequences from the ileum clustered with Env HIV-1 RNA sequences in the jejunum (and plasma) and to a lesser extent the duodenum, suggesting that there was extensive exchange of viral variants between the ileum and other tissue compartments including blood.
Differential distribution of Env variants with altered neutralization and co-receptor binding domains
The primary antibody neutralization domain of subtype C, a GPGQ tetramer located at the tip of the V3 loop, was conserved in 97.0% (197/203) of GIT and 100% (9/9) of plasma samples. Six tetramer motifs, four in patient HIV021 (GPGH in the duodenum; GPGR and 2 RPGQ in the right colon) and two in patient HIV029 (RPGQ in the duodenum; RPGY in the left colon) had a basic amino acid substitution in the first or last position of the tetramer, either arginine (R) or histidine (H). An analysis of tropism using three different algorithms, the Web Position Specific Scoring Matrix (PSSM) tool (SINSI matrix), the Geno2pheno and the CoRSeqv3C co-receptor predictor algorithm indicated that, depending on the algorithm used, from 83.1% to 100% of all variants in the GIT of patients HIV019 (n = 72), HIV021 (n = 66) and HIV029 (n = 65), were CCR5-tropic and non-syncytium inducing (NSI). The most sensitive method, CoRSeqv3C, identified 5 CXCR4-tropic variants in the duodenum (n = 1), left colon (n = 1) and right colon (n = 3) of patient HIV021. These variants included the four viral quasispecies with a basic amino acid substitution in the tetrameric GPGQ tip of the V3 loop and a fifth variant with a basic (arginine) residue at position 1 of the V3 loop. A total of 11 CXCR4-tropic variants were identified in samples obtained from the duodenum (n=1), jejunum (n = 5), ileum (n =1), left colon (n=1) and plasma (n = 3) of patient HIV029 using the CoRSeqv3C method. All of these variants contained a basic substitution in the V3 loop, either a lysine residue at position 32 (n=9) or an arginine residue at position 15 (n=2). No CXCR4-using variants were detected in the GIT of patient HIV019 using any of the 3 testing algorithms.
DISCUSSION
There are still many unanswered questions relating to the role of the GIT in the genetic evolution and dynamics of HIV-1 infection. It has also been difficult to compare the data generated in different studies due to inter-study differences in sampling sites and the stage of HIV-1 disease, as well as the confounding effects of different treatment regimens. In addition, most studies have been performed on HIV-1 proviral DNA and thus, may not be representative of the full spectrum of replication competent viral quasispecies. In this study, we examined the diversity of HIV-1 RNA quasispecies in the intestine of a well-characterized cohort of African patients with AIDS, chronic diarrhea and (or) unexplained weight loss. To our knowledge this is the first study of HIV-1 intestinal diversity in patients infected with subtype C, a viral subtype that may be particularly well adapted to replication in the GIT (Sala et al., 2006; Centlivre et al., 2006). It is also the first study to be conducted in Africa, a region of the world where gastrointestinal symptoms and wasting (Slim’ disease) are common. In our study, a major effort was made to exclude patients with enteric co-infections (stool cultures, histological staining for acid-fast bacilli, fungal and parasitic infections). Despite these efforts, we cannot conclusively rule out the possibility that other co-infectious may have affected our results.
Overall, our results are in agreement with a pre-HAART study of proviral DNA in patients infected with subtype B (van Marle et al., 2007). Of interest, was the higher overall diversity of Env compared to gag HIV-1 RNA variants and the finding that Env variants were, in general, more heterogeneous in the small intestine than the colon. These findings suggest that the gag and Env genes are under selection pressures and that these selection pressures vary in different parts of the GIT. Factors contributing to the HIV-1 diversity in different regions of intestine have not been defined but may, among others, include site-specific differences in inflammation and immune activation and in the magnitude and specificity of the anti-HIV-1 CD8+ CTL response, as well as factors related to differences in CD4+ T cell levels, target cell tropism and viral entry, all of which would be expected to have a greater impact on the Env than the gag gene.
Phylogenetic analysis indicated that there were also distinct differences in the distribution and clustering of gag and Env RNA variants. The extensive migration and mixing of gag HIV-1 RNA variants between plasma and different regions of the GIT is consistent with the view that, in the absence of ART, there is widespread dissemination of newly produced viral particles and (or) infected cells. Env RNA variants exhibited a more complex pattern. In addition to mixed phylogenetic clusters, Env trees contained large monophyletic clusters of closely related RNA variants. The presence of these clusters, especially in the colon, suggests the presence of distinct foci of HIV-1 replication that could contribute to the emergence of new viral variants with altered pathogenicity. Additional longitudinal studies will be required to further confirm that there is isolated generation of pathogenic variants of HIV-1 in the GIT of ART-naïve patients. To be informative, these studies will require sequencing of a highly variable gene region that is under strong selection pressure. The reason for the consistent and preferential evolution of Env RNA variants in the colon is not known but may be related to the higher levels of viral replication and target cell availability in the lower vs. upper GIT (Cassol et al., 2013; Schneider et al., 1995; Yukl et al., 2010). Alternatively, it may be more closed related to microbial translocation and high levels of non-specific inflammation and immune activation in the colon.
An analysis of inferred amino acid sequences indicated that some nucleotide substitutions in the Env gene involved the replacement of neutral or acidic amino acids with positively charged residues that are known to play a role in the switch from CCR5 to CXCR4 co-receptor usage (Jensen et al., 2003). These CXCR4-tropic variants were detected in 2 of 3 patients and were present in some, but not all, parts of the GIT. Although HIV-1 subtype C viruses generally use the CCR5 chemokine receptor (along with CD4) for virus entry into susceptible target cells, genotypic changes can occur that allow C viruses to use CXCR4 as an entry receptor. The switch to CXCR4 usage is associated with accelerated CD4+ T cell depletion and progression to AIDS (Connor et al., 1997). Additional in vitro testing will be required to confirm the tropism of these CXCR4-encoding variants. Using population-based sequencing, a method that is commonly used to screen for HIV-1 resistance mutations in plasma RNA, we also detected a number of nucleotide substitutions and inferred amino acid polymorphisms that have been linked to altered susceptibility to ART. As observed for the Env gene and for primary resistance mutations in patients on mono- or dual-therapy (van Marle et al., 2010) and in ART-treated patients with ongoing viral replication (>1,000 HIV-1 RNA copies/mL of plasma) (poles et al., 2001), these resistance-associated variants were not present in all patients, and when present, were differentially distributed throughout the GIT.
The above results are consistent with other studies conducted in pre-HAART patients (van Marle et al., 2007; van Marle et al., 2010) and in viremic patients on HAART (poles et al., 2001). They underscore the highly dynamic nature of HIV-1 infection and RNA diversification in the GIT of ART-naïve patients and suggest that diversification should be viewed both in terms of genetic evolution and migration into, and out of different compartments in the GIT. Strengths of our study relate to the homogeneity of our study cohort with respect to stage of disease, the focus on RNA rather than proviral DNA and the sampling of multiple tissue sites. Limitations relate to the small sample size and the potential biases introduced by population-based PCR cloning (Taq induced recombination, nucleotide misi-ncorporation) (Meyerhan et al., 1990; Yang et al., 1996). However, this does not appear to be a problem when analyzing samples with a high number of PCR templates, as in the current study (Jordan et al., 2010).
It is important to note that several recent studies conducted in HAART-treated patients with plasma viral loads <50 copies/mL have minimized the role of the GIT in the evolution and compartmentalization of HIV-1 and suggested that the GIT is not a source of rebound virus during therapeutic failure (Imamichi et al., 2011; Lerner et al., 2011; Evering et al., 2012). The reason(s) for the failure to detect HIV-1 diversification in patients on maximally suppressive HAART is not known but may be related to the marked decreased viral replication and de novo infection of new target cells, or to reduced chemokine levels and decreased trafficking of HIV-1-infected cells. Alternatively, virus transmission in patients on highly suppressive ART may occur via a cell-cell route (Sigal et al., 2011) that does not involve exposure to strong diversifying selection pressure. Of interest in this regard, is a study by Avettand-Fenoel et al., showing that, following the introduction of HAART, there is reduced viral replication and trafficking between the rectum and peripheral blood (Avettand-Fenoel et al., 2011).
In conclusion, this finding, together with our data, highlight the need for more systematic and comprehensive studies of HIV-1 diversification in different compartments of the upper and lower GIT, both as a function of disease progression and in response to therapy. In addition to serving as a baseline for improved therapeutic monitoring, such studies may provide new insights for preventing the trafficking, dissemination and evolution of more pathogenic and drug resistant strains of HIV-1. These studies will be challenging given the technical and ethical considerations associated with repeat sampling of multiple sites.
ACKNOWLEDGMENTS
We owe a special thanks to the research nurse and endoscopy nurses of the Unitas and Pretoria East GI-units and to Dr. Mark Theron, the study anesthetist, for their dedication and commitment to patient care. We would also like to thank Justen Manasa, Prevashinee Padayachee and Hloniphile Mthiyane for their assistance and sharing their expertise in cloning and sequencing. Most of all, we thank our patients and volunteers. Without their participation and commitment this study would not have been possible.
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. See comment in PubMed Commons below Science. 1998; 280: 427-431.
- Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. See comment in PubMed Commons below J Virol. 2003; 77: 11708-11717.
- Mehandru S, poles MA, Tenner-Racz K, Horowitz A, Hurley A, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. See comment in PubMed Commons below J Exp Med. 2004; 200: 761-770.
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. See comment in PubMed Commons below J Exp Med. 2004; 200: 749-759.
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, et al. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. See comment in PubMed Commons below Nature. 2005; 434: 1093-1097.
- Guadalupe M, Sankaran S, George MD, Reay E, Verhoeven D, et al. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. See comment in PubMed Commons below J Virol. 2006; 80: 8236-8247.
- Centlivre M, Sala M, Wain-Hobson S, Berkhout B. In HIV-1 pathogenesis the die is cast during primary infection. See comment in PubMed Commons below AIDS. 2007; 21: 1-11.
- Brenchley JM, Price DA, Douek DC. HIV disease: fallout from a mucosal catastrophe? See comment in PubMed Commons below Nat Immunol. 2006; 7: 235-239.
- Paiardini M, Frank I, Pandrea I, Apetrei C, Silvestri G. Mucosal immune dysfunction in AIDS pathogenesis. See comment in PubMed Commons below AIDS Rev. 2008; 10: 36-46.
- Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. See comment in PubMed Commons below Mucosal Immunol. 2008; 1: 23-30.
- Mehandru S, poles MA, Tenner-Racz K, Jean-Pierre P, Manuelli V, Lopez P, Shet A . Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. See comment in PubMed Commons below PLoS Med. 2006; 3: e484.
- poles MA, Boscardin WJ, Elliott J, Taing P, Fuerst MM, et al. Lack of decay of HIV-1 in gut-associated lymphoid tissue reservoirs in maximally suppressed individuals. See comment in PubMed Commons below J Acquir Immune Defic Syndr. 2006; 43: 65-68.
- Cassol E, Malfeld S, Mahasha P, van der Merwe S, Cassol S, et al. Persistent microbial translocation and immune activation in HIV-1-infected South Africans receiving combination antiretroviral therapy. See comment in PubMed Commons below J Infect Dis. 2010; 202: 723-733.
- poles MA, Elliott J, Vingerhoets J, Michiels L, Scholliers A. (2001) Despite high concordance, distinct mutational and phenotypic drug resistance profiles in human immunodeficiency virus type 1RNA are observed in gastrointestinal mucosal biopsy specimens and peripheral blood mononuclear cells compared with plasma. J Infect Dis 183:143-8.
- Avettand-Fenoel V, Hocqueloux L, Müller-Trutwin M, Prazuck T, Melard A, et al. Greater diversity of HIV DNA variants in the rectum compared to variants in the blood in patients without HAART. See comment in PubMed Commons below J Med Virol. 2011; 83: 1499-1507.
- Imamichi H, Degray G, Dewar RL, Mannon P, Yao M, et al. Lack of compartmentalization of HIV-1 quasispecies between the gut and peripheral blood compartments. See comment in PubMed Commons below J Infect Dis. 2011; 204: 309-314.
- Lerner P, Guadalupe M, Donovan R, Hung J, Flamm J, et al. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. See comment in PubMed Commons below J Virol. 2011; 85: 4772-4782.
- Evering TH, Mehandru S, Racz P, Tenner-Racz K, poles MA, et al. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. See comment in PubMed Commons below PLoS Pathog. 2012; 8: e1002506.
- Cassol E, Malfeld S, Mahasha P, Bond R, Slavik T, et al. Impaired CD4+ T-cell restoration in the small versus large intestine of HIV-1-positive South Africans receiving combination antiretroviral therapy. See comment in PubMed Commons below J Infect Dis. 2013; 208: 1113-1122.
- Van Laethem K, Schrooten Y, Dedecker S, Van Heeswijck L, Deforche K, et al. A genotypic assay for the amplification and sequencing of gag and protease from diverse human immunodeficiency virus type 1 group M subtypes. See comment in PubMed Commons below J Virol Methods. 2006; 132: 181-186.
- Pillay V (2006) In-house assay for pol sequencing to monitor ARV Drug Resistance National Health Laboratory Service (South Africa), Q-Pulse4/docs/active/ NIC0085: 1-12.
- Gordon M, De Oliveira T, Bishop K, Coovadia HM, Madurai L, et al. Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies. See comment in PubMed Commons below J Virol. 2003; 77: 2587-2599.
- Rousseau CM, Birditt BA, McKay AR, Stoddard JN, Lee TC, et al. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. See comment in PubMed Commons below J Virol Methods. 2006; 136: 118-125.
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acid Res 22: 4673-4680.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. See comment in PubMed Commons below J Mol Evol. 1980; 16: 111-120.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. See comment in PubMed Commons below Mol Biol Evol. 2011; 28: 2731-2739.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. See comment in PubMed Commons below Syst Biol. 2010; 59: 307-321.
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. See comment in PubMed Commons below Bioinformatics. 2001; 17: 754-755.
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. See comment in PubMed Commons below Bioinformatics. 2003; 19: 1572-1574.
- Drummond A, Rambaut A. Tracer [Online]. Website last modified on February 9, 2008 (accessed on August 18, 2011). Available at http://beast.bio.ed.ac.uk/ Tracer
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. See comment in PubMed Commons below Mol Biol Evol. 2012; 29: 1969-1973.
- Jensen MA, Li FS, van't Wout AB, Nickle DC, Shriner D. (2003) Improved co-receptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 Env V3 loop sequences. J Virol 77:13376-13388. Available at http://indra.mullins.microbiol.washington. edu/webpssm/.
- Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R . Bioinformatics prediction of HIV coreceptor usage. See comment in PubMed Commons below Nat Biotechnol. 2007; 25: 1407-1410.
- Cashin K, Gray LR, Jakobsen MR, Sterjovski J, Churchill MJ, et al. (2013) CoRSeqV3-C: a novel HIV-1 subtype C specific V3 sequence based coreceptor usage prediction algorithm. Retrovirol 10:1-10. Available online at http:// tools.burnet.edu.au/coreceptor/
- Velazquez-Campoy A, Todd MJ, Vega S, Freire E. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. See comment in PubMed Commons below Proc Natl Acad Sci U S A. 2001; 98: 6062-6067.
- Gonzalez LMF, Brindeiro RM, Tarin M, Calazans A, Soares MA. (2003) The subtype C of human immunodeficiency virus type 1 protease presents an in vitro hypersusceptibility to the protease inhibitor Lopinavir. Antimicrob. Agents Chemother 47:2817-2822.
- Sala M, Centlivre M, Wain-Hobson S. Clade-specific differences in active viral replication and compartmentalization. See comment in PubMed Commons below Curr Opin HIV AIDS. 2006; 1: 108-114.
- Centlivre M, Sommer P, Michel M, Ho Tsong Fang R, Gofflo S, et al. The HIV-1 clade C promoter is particularly well adapted to replication in the gut in primary infection. See comment in PubMed Commons below AIDS. 2006; 20: 657-666.
- van Marle G, Gill MJ, Kolodka D, McManus L, Grant T, et al. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. See comment in PubMed Commons below Retrovirology. 2007; 4: 87.
- Schneider T, Jahn HU, Schmidt W, Riecken EO, Zeitz M, et al. Loss of CD4 T lymphocytes in patients infected with human immunodeficiency virus type 1 is more pronounced in the duodenal mucosa than in the peripheral blood. Berlin Diarrhea/Wasting Syndrome Study Group. See comment in PubMed Commons below Gut. 1995; 37: 524-529.
- Yukl SA, Gianella S, Sinclair E, Epling L, Li Q, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. See comment in PubMed Commons below J Infect Dis. 2010; 202: 1553-1561.
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. See comment in PubMed Commons below J Exp Med. 1997; 185: 621-628.
- van Marle G, Church DL, Nunweiler KD, Cannon K, Wainberg MA, et al. Higher levels of Zidovudine resistant HIV in the colon compared to blood and other gastrointestinal compartments in HIV infection. See comment in PubMed Commons below Retrovirology. 2010; 7: 74.
- Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. See comment in PubMed Commons below Nucleic Acids Res. 1990; 18: 1687-1691.
- Yang YL, Wang G, Dorman K, Kaplan AH. Long polymerase chain reaction amplification of heterogeneous HIV type 1 templates produces recombination at a relatively high frequency. See comment in PubMed Commons below AIDS Res Hum Retroviruses. 1996; 12: 303-306.
- Jordan MR, Kearney M, Palmer S, Shao W, Maldarelli F, et al. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. See comment in PubMed Commons below J Virol Methods. 2010; 168: 114-120.
- Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, et al. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. See comment in PubMed Commons below Nature. 2011; 477: 95-98.


Sign up for Article Alerts